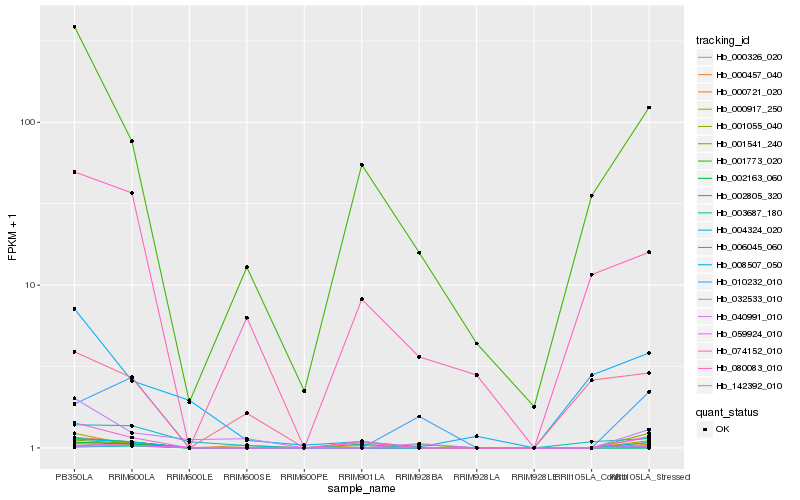
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006045_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104222982, partial [Nicotiana sylvestris] |
| 2 |
Hb_000721_020 |
0.009149328 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 3 |
Hb_000326_020 |
0.1196671414 |
- |
- |
- |
| 4 |
Hb_032533_010 |
0.2123573063 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 5 |
Hb_003687_180 |
0.2234937809 |
- |
- |
hypothetical protein JCGZ_02335 [Jatropha curcas] |
| 6 |
Hb_074152_010 |
0.2418474075 |
- |
- |
- |
| 7 |
Hb_000917_250 |
0.2907209152 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 130-like [Populus euphratica] |
| 8 |
Hb_001055_040 |
0.3127169095 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 9 |
Hb_010232_010 |
0.3227382819 |
- |
- |
- |
| 10 |
Hb_059924_010 |
0.3246521208 |
- |
- |
- |
| 11 |
Hb_001541_240 |
0.3432007221 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_040991_010 |
0.3535725616 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X2 [Jatropha curcas] |
| 13 |
Hb_001773_020 |
0.3598858189 |
- |
- |
hypothetical protein POPTR_0016s00980g [Populus trichocarpa] |
| 14 |
Hb_080083_010 |
0.3625287343 |
- |
- |
- |
| 15 |
Hb_000457_040 |
0.3707138465 |
- |
- |
- |
| 16 |
Hb_004324_020 |
0.3757054453 |
- |
- |
- |
| 17 |
Hb_008507_050 |
0.3845392862 |
- |
- |
PREDICTED: S-adenosylmethionine decarboxylase proenzyme-like [Jatropha curcas] |
| 18 |
Hb_142392_010 |
0.3848297353 |
- |
- |
PREDICTED: uncharacterized protein LOC105628741 [Jatropha curcas] |
| 19 |
Hb_002163_060 |
0.3850231817 |
- |
- |
- |
| 20 |
Hb_002805_320 |
0.3850519129 |
- |
- |
- |