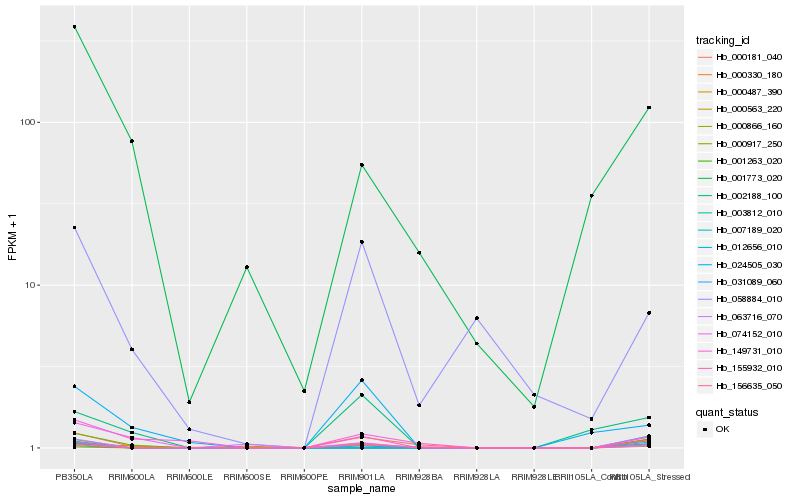
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007189_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 2 |
Hb_000330_180 |
0.197615954 |
- |
- |
- |
| 3 |
Hb_000917_250 |
0.262600224 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 130-like [Populus euphratica] |
| 4 |
Hb_000181_040 |
0.2739111926 |
transcription factor |
TF Family: NAC |
PREDICTED: protein CUP-SHAPED COTYLEDON 2 [Jatropha curcas] |
| 5 |
Hb_155932_010 |
0.330931147 |
- |
- |
PREDICTED: uncharacterized protein LOC105786773 [Gossypium raimondii] |
| 6 |
Hb_031089_060 |
0.337415329 |
- |
- |
- |
| 7 |
Hb_000866_160 |
0.3377514557 |
- |
- |
PREDICTED: elicitor-responsive protein 3 [Populus euphratica] |
| 8 |
Hb_074152_010 |
0.3409280284 |
- |
- |
- |
| 9 |
Hb_156635_050 |
0.3470557538 |
- |
- |
hypothetical protein JCGZ_01613 [Jatropha curcas] |
| 10 |
Hb_001773_020 |
0.371361748 |
- |
- |
hypothetical protein POPTR_0016s00980g [Populus trichocarpa] |
| 11 |
Hb_000487_390 |
0.3858690885 |
- |
- |
PREDICTED: putative pumilio homolog 21 [Jatropha curcas] |
| 12 |
Hb_001263_020 |
0.3882487172 |
- |
- |
- |
| 13 |
Hb_012656_010 |
0.3888462698 |
- |
- |
- |
| 14 |
Hb_058884_010 |
0.3902760378 |
- |
- |
PREDICTED: barwin-like [Jatropha curcas] |
| 15 |
Hb_063716_070 |
0.392582259 |
- |
- |
hypothetical protein PRUPE_ppa020565mg [Prunus persica] |
| 16 |
Hb_003812_010 |
0.3937359147 |
- |
- |
- |
| 17 |
Hb_000563_220 |
0.3952678288 |
- |
- |
hypothetical protein VITISV_004111 [Vitis vinifera] |
| 18 |
Hb_149731_010 |
0.3973686305 |
- |
- |
- |
| 19 |
Hb_024505_030 |
0.3992607783 |
- |
- |
PREDICTED: aspartate aminotransferase, chloroplastic [Jatropha curcas] |
| 20 |
Hb_002188_100 |
0.4005202668 |
- |
- |
conserved hypothetical protein [Ricinus communis] |