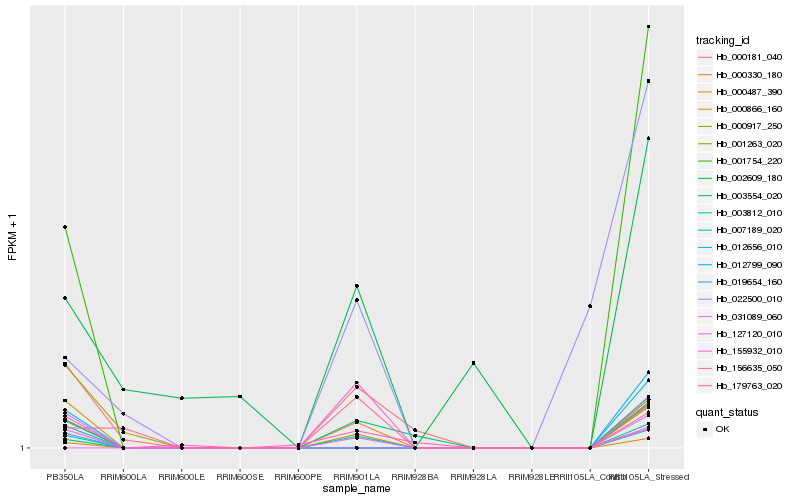
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000330_180 |
0.0 |
- |
- |
- |
| 2 |
Hb_007189_020 |
0.197615954 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 3 |
Hb_156635_050 |
0.2559191113 |
- |
- |
hypothetical protein JCGZ_01613 [Jatropha curcas] |
| 4 |
Hb_003554_020 |
0.3169159531 |
- |
- |
hypothetical protein JCGZ_14833 [Jatropha curcas] |
| 5 |
Hb_155932_010 |
0.338535573 |
- |
- |
PREDICTED: uncharacterized protein LOC105786773 [Gossypium raimondii] |
| 6 |
Hb_000181_040 |
0.341817241 |
transcription factor |
TF Family: NAC |
PREDICTED: protein CUP-SHAPED COTYLEDON 2 [Jatropha curcas] |
| 7 |
Hb_179763_020 |
0.3538808886 |
- |
- |
- |
| 8 |
Hb_001263_020 |
0.3632704409 |
- |
- |
- |
| 9 |
Hb_012656_010 |
0.3632715226 |
- |
- |
- |
| 10 |
Hb_000487_390 |
0.3633010331 |
- |
- |
PREDICTED: putative pumilio homolog 21 [Jatropha curcas] |
| 11 |
Hb_003812_010 |
0.3634063542 |
- |
- |
- |
| 12 |
Hb_022500_010 |
0.3646820438 |
- |
- |
- |
| 13 |
Hb_002609_180 |
0.3653674277 |
- |
- |
- |
| 14 |
Hb_001754_220 |
0.3680555274 |
- |
- |
- |
| 15 |
Hb_000917_250 |
0.3731878941 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 130-like [Populus euphratica] |
| 16 |
Hb_019654_160 |
0.3825745234 |
- |
- |
Zinc finger protein GIS2 [Medicago truncatula] |
| 17 |
Hb_012799_090 |
0.3829989828 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g29230-like [Jatropha curcas] |
| 18 |
Hb_000866_160 |
0.3852698534 |
- |
- |
PREDICTED: elicitor-responsive protein 3 [Populus euphratica] |
| 19 |
Hb_031089_060 |
0.3857590744 |
- |
- |
- |
| 20 |
Hb_127120_010 |
0.3861150042 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |