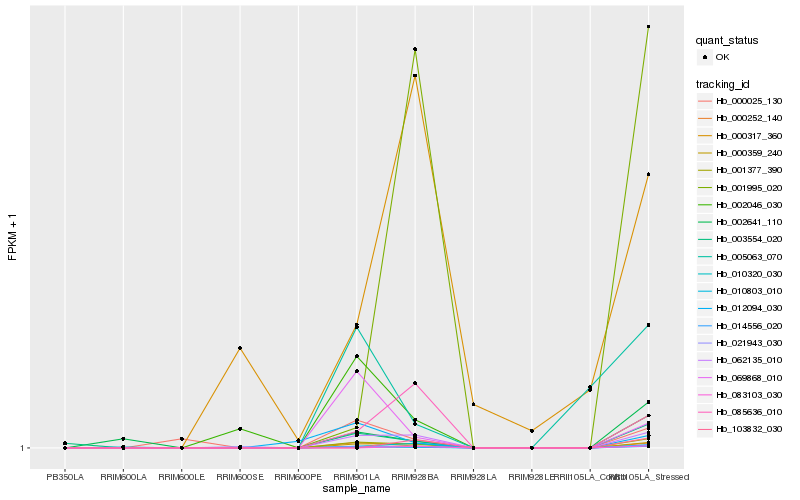
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000359_240 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105646039 [Jatropha curcas] |
| 2 |
Hb_062135_010 |
0.0127717884 |
- |
- |
hypothetical protein POPTR_0011s036602g, partial [Populus trichocarpa] |
| 3 |
Hb_001377_390 |
0.1162893516 |
- |
- |
hypothetical protein POPTR_0002s17390g [Populus trichocarpa] |
| 4 |
Hb_014556_020 |
0.1196401264 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 5 |
Hb_000252_140 |
0.1731266884 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 6 |
Hb_010320_030 |
0.1775440067 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 7 |
Hb_085636_010 |
0.2450231258 |
- |
- |
PREDICTED: uncharacterized protein LOC105636643 [Jatropha curcas] |
| 8 |
Hb_003554_020 |
0.2768925425 |
- |
- |
hypothetical protein JCGZ_14833 [Jatropha curcas] |
| 9 |
Hb_021943_030 |
0.3285136354 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 10 |
Hb_000025_130 |
0.3353247043 |
- |
- |
- |
| 11 |
Hb_002641_110 |
0.3603477201 |
- |
- |
PREDICTED: uncharacterized protein LOC103448574 [Malus domestica] |
| 12 |
Hb_000317_360 |
0.3663224649 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g08460 [Jatropha curcas] |
| 13 |
Hb_002046_030 |
0.3694886886 |
- |
- |
PREDICTED: uncharacterized protein LOC104788746 [Camelina sativa] |
| 14 |
Hb_010803_010 |
0.37037328 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 15 |
Hb_069868_010 |
0.372233011 |
- |
- |
hypothetical protein JCGZ_14833 [Jatropha curcas] |
| 16 |
Hb_001995_020 |
0.3723410701 |
- |
- |
PREDICTED: trans-resveratrol di-O-methyltransferase-like [Populus euphratica] |
| 17 |
Hb_012094_030 |
0.3786712815 |
- |
- |
PREDICTED: uncharacterized protein LOC103699727 [Phoenix dactylifera] |
| 18 |
Hb_005063_070 |
0.3947783215 |
- |
- |
PREDICTED: PRA1 family protein E [Jatropha curcas] |
| 19 |
Hb_083103_030 |
0.403853774 |
- |
- |
putative retrotransposon protein [Malus domestica] |
| 20 |
Hb_103832_030 |
0.4038662488 |
- |
- |
PREDICTED: uncharacterized protein LOC105795045 [Gossypium raimondii] |