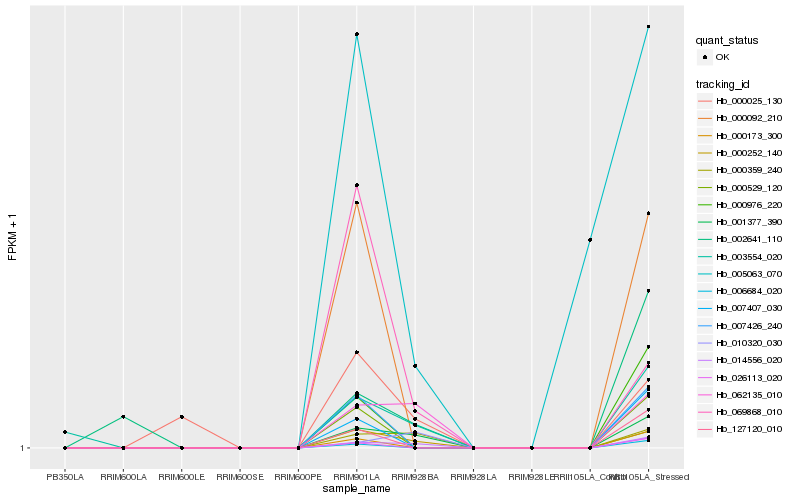
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001377_390 |
0.0 |
- |
- |
hypothetical protein POPTR_0002s17390g [Populus trichocarpa] |
| 2 |
Hb_014556_020 |
0.0326470936 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 3 |
Hb_000252_140 |
0.1162599787 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_000359_240 |
0.1162893516 |
- |
- |
PREDICTED: uncharacterized protein LOC105646039 [Jatropha curcas] |
| 5 |
Hb_062135_010 |
0.1199786091 |
- |
- |
hypothetical protein POPTR_0011s036602g, partial [Populus trichocarpa] |
| 6 |
Hb_003554_020 |
0.2243282649 |
- |
- |
hypothetical protein JCGZ_14833 [Jatropha curcas] |
| 7 |
Hb_010320_030 |
0.2817886947 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 8 |
Hb_002641_110 |
0.2938970369 |
- |
- |
PREDICTED: uncharacterized protein LOC103448574 [Malus domestica] |
| 9 |
Hb_000025_130 |
0.3121592542 |
- |
- |
- |
| 10 |
Hb_000173_300 |
0.3281274387 |
- |
- |
PREDICTED: uncharacterized protein LOC105631894 [Jatropha curcas] |
| 11 |
Hb_026113_020 |
0.3290506561 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 12 |
Hb_006684_020 |
0.3293799668 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 13 |
Hb_000529_120 |
0.3297657705 |
- |
- |
PREDICTED: uncharacterized protein LOC105641745 [Jatropha curcas] |
| 14 |
Hb_000976_220 |
0.3307257191 |
- |
- |
PREDICTED: protein ALTERED XYLOGLUCAN 4-like [Jatropha curcas] |
| 15 |
Hb_127120_010 |
0.330733074 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 16 |
Hb_007407_030 |
0.330760317 |
- |
- |
hypothetical protein JCGZ_23510 [Jatropha curcas] |
| 17 |
Hb_007426_240 |
0.3329883781 |
- |
- |
- |
| 18 |
Hb_000092_210 |
0.3429077607 |
- |
- |
- |
| 19 |
Hb_005063_070 |
0.3437245381 |
- |
- |
PREDICTED: PRA1 family protein E [Jatropha curcas] |
| 20 |
Hb_069868_010 |
0.3461921431 |
- |
- |
hypothetical protein JCGZ_14833 [Jatropha curcas] |