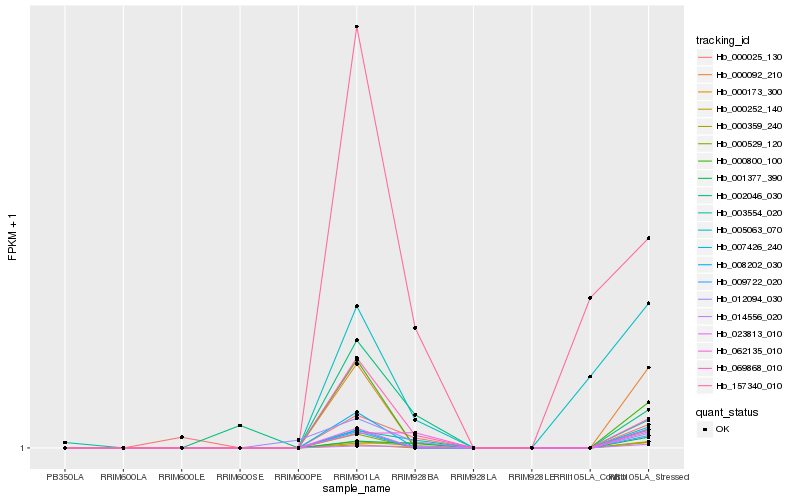
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000252_140 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_001377_390 |
0.1162599787 |
- |
- |
hypothetical protein POPTR_0002s17390g [Populus trichocarpa] |
| 3 |
Hb_014556_020 |
0.148579355 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 4 |
Hb_062135_010 |
0.1675642569 |
- |
- |
hypothetical protein POPTR_0011s036602g, partial [Populus trichocarpa] |
| 5 |
Hb_000359_240 |
0.1731266884 |
- |
- |
PREDICTED: uncharacterized protein LOC105646039 [Jatropha curcas] |
| 6 |
Hb_069868_010 |
0.2332466003 |
- |
- |
hypothetical protein JCGZ_14833 [Jatropha curcas] |
| 7 |
Hb_003554_020 |
0.2441268982 |
- |
- |
hypothetical protein JCGZ_14833 [Jatropha curcas] |
| 8 |
Hb_000025_130 |
0.2718002942 |
- |
- |
- |
| 9 |
Hb_000092_210 |
0.2909550742 |
- |
- |
- |
| 10 |
Hb_023813_010 |
0.2915032605 |
- |
- |
PREDICTED: uncharacterized protein LOC105162193 [Sesamum indicum] |
| 11 |
Hb_012094_030 |
0.2927640788 |
- |
- |
PREDICTED: uncharacterized protein LOC103699727 [Phoenix dactylifera] |
| 12 |
Hb_009722_020 |
0.2928237499 |
- |
- |
- |
| 13 |
Hb_007426_240 |
0.2952699538 |
- |
- |
- |
| 14 |
Hb_002046_030 |
0.2977191611 |
- |
- |
PREDICTED: uncharacterized protein LOC104788746 [Camelina sativa] |
| 15 |
Hb_000529_120 |
0.2996374696 |
- |
- |
PREDICTED: uncharacterized protein LOC105641745 [Jatropha curcas] |
| 16 |
Hb_008202_030 |
0.311881759 |
- |
- |
hypothetical protein B456_002G115600 [Gossypium raimondii] |
| 17 |
Hb_000800_100 |
0.3171906755 |
- |
- |
hypothetical protein JCGZ_01543 [Jatropha curcas] |
| 18 |
Hb_005063_070 |
0.3184277148 |
- |
- |
PREDICTED: PRA1 family protein E [Jatropha curcas] |
| 19 |
Hb_000173_300 |
0.3221893783 |
- |
- |
PREDICTED: uncharacterized protein LOC105631894 [Jatropha curcas] |
| 20 |
Hb_157340_010 |
0.3236328944 |
- |
- |
hypothetical protein CISIN_1g0109951mg, partial [Citrus sinensis] |