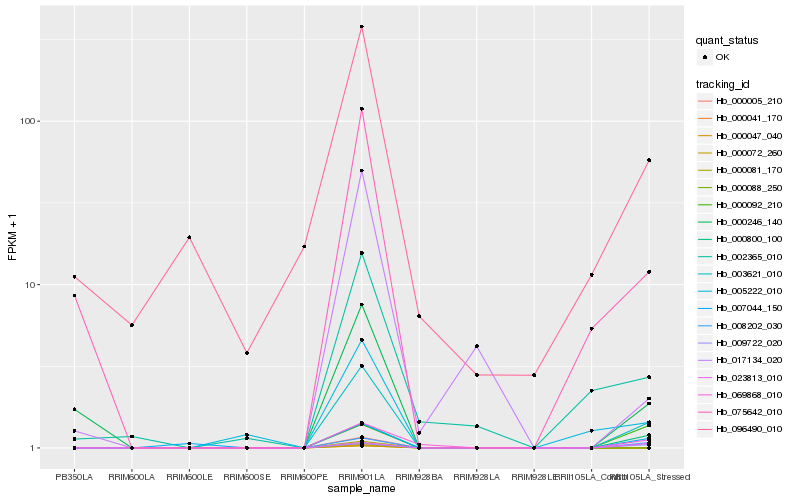
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003621_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_000800_100 |
0.1558531875 |
- |
- |
hypothetical protein JCGZ_01543 [Jatropha curcas] |
| 3 |
Hb_008202_030 |
0.170295285 |
- |
- |
hypothetical protein B456_002G115600 [Gossypium raimondii] |
| 4 |
Hb_069868_010 |
0.2182086825 |
- |
- |
hypothetical protein JCGZ_14833 [Jatropha curcas] |
| 5 |
Hb_000246_140 |
0.2219933806 |
- |
- |
xylulose kinase, putative [Ricinus communis] |
| 6 |
Hb_075642_010 |
0.2325301197 |
- |
- |
PREDICTED: uncharacterized protein LOC105647831 [Jatropha curcas] |
| 7 |
Hb_005222_010 |
0.2463309262 |
- |
- |
hypothetical protein MTR_1g033950 [Medicago truncatula] |
| 8 |
Hb_009722_020 |
0.2506077521 |
- |
- |
- |
| 9 |
Hb_007044_150 |
0.2534173184 |
- |
- |
PREDICTED: DNA-directed RNA polymerases II and V subunit 8A-like [Jatropha curcas] |
| 10 |
Hb_023813_010 |
0.2649147048 |
- |
- |
PREDICTED: uncharacterized protein LOC105162193 [Sesamum indicum] |
| 11 |
Hb_002365_010 |
0.2743209977 |
- |
- |
hypothetical protein CISIN_1g0405342mg, partial [Citrus sinensis] |
| 12 |
Hb_096490_010 |
0.2760907941 |
- |
- |
hypothetical protein CICLE_v100210772mg, partial [Citrus clementina] |
| 13 |
Hb_017134_020 |
0.2903231053 |
- |
- |
- |
| 14 |
Hb_000092_210 |
0.2955100629 |
- |
- |
- |
| 15 |
Hb_000005_210 |
0.2971839715 |
- |
- |
- |
| 16 |
Hb_000041_170 |
0.2971839715 |
- |
- |
PREDICTED: uncharacterized protein At4g02000-like [Eucalyptus grandis] |
| 17 |
Hb_000047_040 |
0.2971839715 |
- |
- |
PREDICTED: uncharacterized protein LOC105775709 [Gossypium raimondii] |
| 18 |
Hb_000072_260 |
0.2971839715 |
- |
- |
hypothetical protein MTR_5g044090 [Medicago truncatula] |
| 19 |
Hb_000081_170 |
0.2971839715 |
- |
- |
- |
| 20 |
Hb_000088_250 |
0.2971839715 |
- |
- |
- |