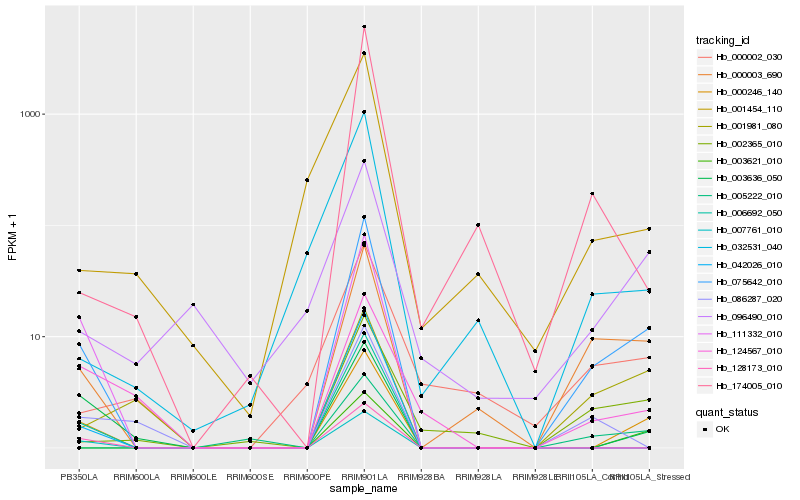
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_075642_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647831 [Jatropha curcas] |
| 2 |
Hb_000246_140 |
0.1432438044 |
- |
- |
xylulose kinase, putative [Ricinus communis] |
| 3 |
Hb_000003_690 |
0.1511586707 |
- |
- |
- |
| 4 |
Hb_002365_010 |
0.2247072059 |
- |
- |
hypothetical protein CISIN_1g0405342mg, partial [Citrus sinensis] |
| 5 |
Hb_003621_010 |
0.2325301197 |
- |
- |
- |
| 6 |
Hb_005222_010 |
0.2335897585 |
- |
- |
hypothetical protein MTR_1g033950 [Medicago truncatula] |
| 7 |
Hb_006692_050 |
0.2378596215 |
- |
- |
Chloroplastic lipocalin [Theobroma cacao] |
| 8 |
Hb_042026_010 |
0.2431131989 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 9 |
Hb_003636_050 |
0.2456246652 |
- |
- |
PREDICTED: bark storage protein A-like [Populus euphratica] |
| 10 |
Hb_096490_010 |
0.2466577801 |
- |
- |
hypothetical protein CICLE_v100210772mg, partial [Citrus clementina] |
| 11 |
Hb_174005_010 |
0.2467765702 |
- |
- |
hypothetical protein EUGRSUZ_D00590 [Eucalyptus grandis] |
| 12 |
Hb_032531_040 |
0.2507655056 |
- |
- |
E3 ISG15--protein ligase HERC5 [Gossypium arboreum] |
| 13 |
Hb_001981_080 |
0.253536069 |
- |
- |
PREDICTED: mitochondrial tRNA-specific 2-thiouridylase 1 isoform X4 [Musa acuminata subsp. malaccensis] |
| 14 |
Hb_007761_010 |
0.2538586927 |
- |
- |
- |
| 15 |
Hb_128173_010 |
0.2547822798 |
- |
- |
PREDICTED: uncharacterized protein LOC104219073 [Nicotiana sylvestris] |
| 16 |
Hb_001454_110 |
0.2615243009 |
- |
- |
hypothetical protein POPTR_0026s00250g [Populus trichocarpa] |
| 17 |
Hb_086287_020 |
0.262602922 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g03880, mitochondrial [Jatropha curcas] |
| 18 |
Hb_000002_030 |
0.2632869526 |
- |
- |
- |
| 19 |
Hb_124567_010 |
0.2637138473 |
- |
- |
- |
| 20 |
Hb_111332_010 |
0.2648277552 |
- |
- |
Dihydroneopterin aldolase [Morus notabilis] |