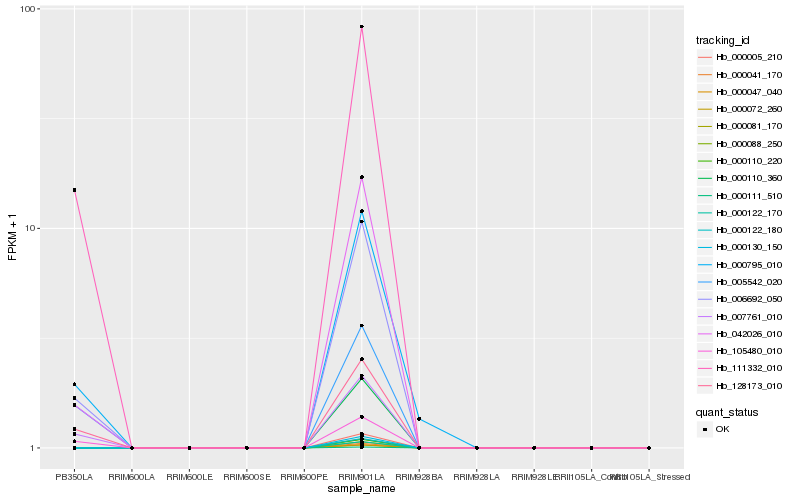
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006692_050 |
0.0 |
- |
- |
Chloroplastic lipocalin [Theobroma cacao] |
| 2 |
Hb_042026_010 |
0.0621876864 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 3 |
Hb_007761_010 |
0.0825507344 |
- |
- |
- |
| 4 |
Hb_128173_010 |
0.0852460016 |
- |
- |
PREDICTED: uncharacterized protein LOC104219073 [Nicotiana sylvestris] |
| 5 |
Hb_111332_010 |
0.1112468218 |
- |
- |
Dihydroneopterin aldolase [Morus notabilis] |
| 6 |
Hb_000795_010 |
0.122981479 |
- |
- |
- |
| 7 |
Hb_105480_010 |
0.1247798894 |
- |
- |
- |
| 8 |
Hb_005542_020 |
0.1480133409 |
- |
- |
lupus la ribonucleoprotein, putative [Ricinus communis] |
| 9 |
Hb_000005_210 |
0.1834934437 |
- |
- |
- |
| 10 |
Hb_000041_170 |
0.1834934437 |
- |
- |
PREDICTED: uncharacterized protein At4g02000-like [Eucalyptus grandis] |
| 11 |
Hb_000047_040 |
0.1834934437 |
- |
- |
PREDICTED: uncharacterized protein LOC105775709 [Gossypium raimondii] |
| 12 |
Hb_000072_260 |
0.1834934437 |
- |
- |
hypothetical protein MTR_5g044090 [Medicago truncatula] |
| 13 |
Hb_000081_170 |
0.1834934437 |
- |
- |
- |
| 14 |
Hb_000088_250 |
0.1834934437 |
- |
- |
- |
| 15 |
Hb_000110_220 |
0.1834934437 |
- |
- |
- |
| 16 |
Hb_000110_360 |
0.1834934437 |
- |
- |
- |
| 17 |
Hb_000111_510 |
0.1834934437 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 18 |
Hb_000122_170 |
0.1834934437 |
- |
- |
PREDICTED: uncharacterized protein LOC104455487 [Eucalyptus grandis] |
| 19 |
Hb_000122_180 |
0.1834934437 |
- |
- |
- |
| 20 |
Hb_000130_150 |
0.1834934437 |
- |
- |
- |