| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
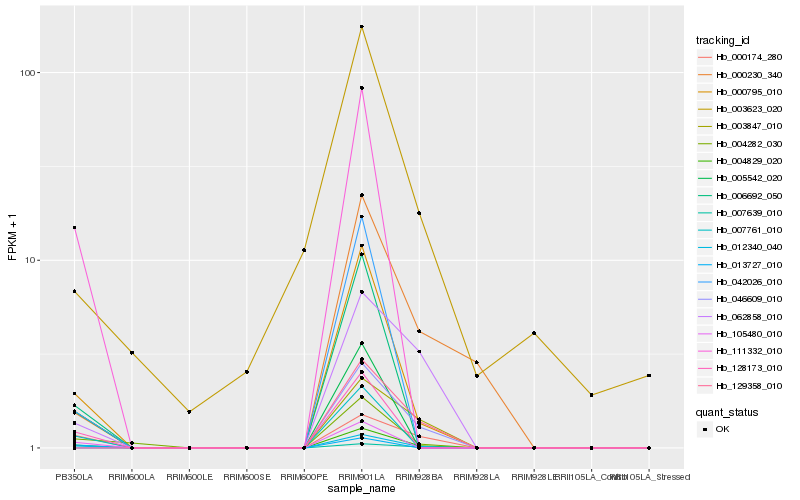
Hb_004829_020 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_012340_040 |
0.0079131879 |
- |
- |
PREDICTED: omega-hydroxypalmitate O-feruloyl transferase-like [Populus euphratica] |
| 3 |
Hb_000795_010 |
0.1092447387 |
- |
- |
- |
| 4 |
Hb_004282_030 |
0.1800079217 |
- |
- |
hypothetical protein EUGRSUZ_B01857 [Eucalyptus grandis] |
| 5 |
Hb_000174_280 |
0.1920663135 |
- |
- |
- |
| 6 |
Hb_007761_010 |
0.2035498398 |
- |
- |
- |
| 7 |
Hb_128173_010 |
0.2036959831 |
- |
- |
PREDICTED: uncharacterized protein LOC104219073 [Nicotiana sylvestris] |
| 8 |
Hb_111332_010 |
0.2068805571 |
- |
- |
Dihydroneopterin aldolase [Morus notabilis] |
| 9 |
Hb_105480_010 |
0.209781771 |
- |
- |
- |
| 10 |
Hb_006692_050 |
0.2154156227 |
- |
- |
Chloroplastic lipocalin [Theobroma cacao] |
| 11 |
Hb_005542_020 |
0.2166376159 |
- |
- |
lupus la ribonucleoprotein, putative [Ricinus communis] |
| 12 |
Hb_003847_010 |
0.2193997062 |
- |
- |
- |
| 13 |
Hb_062858_010 |
0.229965377 |
- |
- |
heat-shock protein, putative [Ricinus communis] |
| 14 |
Hb_013727_010 |
0.2355099276 |
- |
- |
hypothetical protein JCGZ_08656 [Jatropha curcas] |
| 15 |
Hb_042026_010 |
0.2418733545 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 16 |
Hb_046609_010 |
0.2420175955 |
- |
- |
- |
| 17 |
Hb_000230_340 |
0.2453693537 |
- |
- |
Transcription elongation factor, putative [Ricinus communis] |
| 18 |
Hb_129358_010 |
0.2512384606 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 11-like [Jatropha curcas] |
| 19 |
Hb_003623_020 |
0.2593826549 |
- |
- |
- |
| 20 |
Hb_007639_010 |
0.2616044911 |
- |
- |
hypothetical protein JCGZ_11284 [Jatropha curcas] |