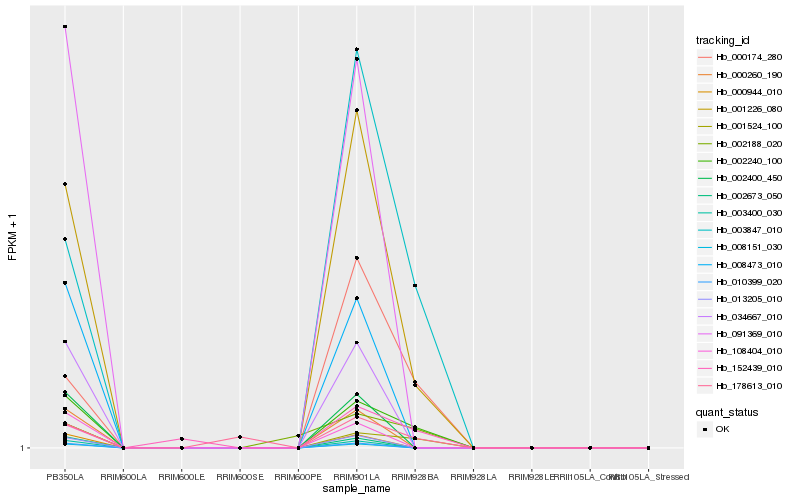
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002240_100 |
0.0 |
- |
- |
- |
| 2 |
Hb_001524_100 |
0.0749306614 |
- |
- |
PREDICTED: uncharacterized protein LOC105635099 [Jatropha curcas] |
| 3 |
Hb_001226_080 |
0.1534093966 |
- |
- |
heat-shock protein, putative [Ricinus communis] |
| 4 |
Hb_003847_010 |
0.1851431852 |
- |
- |
- |
| 5 |
Hb_000174_280 |
0.2243935723 |
- |
- |
- |
| 6 |
Hb_152439_010 |
0.2643527583 |
- |
- |
PREDICTED: uncharacterized protein LOC105796032 [Gossypium raimondii] |
| 7 |
Hb_178613_010 |
0.2842644559 |
- |
- |
PREDICTED: uncharacterized protein LOC105796032 [Gossypium raimondii] |
| 8 |
Hb_002188_020 |
0.2887444852 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 9 |
Hb_008473_010 |
0.2987105701 |
- |
- |
PREDICTED: laccase-14-like [Jatropha curcas] |
| 10 |
Hb_091369_010 |
0.298711735 |
- |
- |
- |
| 11 |
Hb_002673_050 |
0.2987234499 |
- |
- |
- |
| 12 |
Hb_000944_010 |
0.2988872852 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 13 |
Hb_003400_030 |
0.298890521 |
- |
- |
PREDICTED: uncharacterized protein LOC104234330 [Nicotiana sylvestris] |
| 14 |
Hb_002400_450 |
0.2990807518 |
- |
- |
PREDICTED: protein MEMO1 isoform X2 [Sesamum indicum] |
| 15 |
Hb_000260_190 |
0.2992055757 |
- |
- |
- |
| 16 |
Hb_034667_010 |
0.299459689 |
- |
- |
- |
| 17 |
Hb_008151_030 |
0.2999614274 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 18 |
Hb_010399_020 |
0.3003237114 |
- |
- |
PREDICTED: uncharacterized protein LOC104094989 [Nicotiana tomentosiformis] |
| 19 |
Hb_108404_010 |
0.3027005956 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 20 |
Hb_013205_010 |
0.3050653429 |
- |
- |
PREDICTED: putative B3 domain-containing protein At2g27410 [Jatropha curcas] |