| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
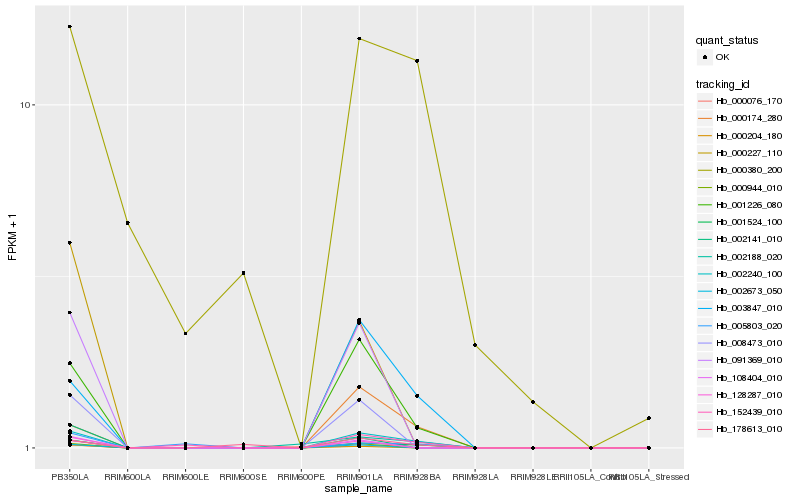
Hb_001524_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635099 [Jatropha curcas] |
| 2 |
Hb_002240_100 |
0.0749306614 |
- |
- |
- |
| 3 |
Hb_001226_080 |
0.2189279133 |
- |
- |
heat-shock protein, putative [Ricinus communis] |
| 4 |
Hb_003847_010 |
0.2531258506 |
- |
- |
- |
| 5 |
Hb_000174_280 |
0.2908050989 |
- |
- |
- |
| 6 |
Hb_005803_020 |
0.3002195348 |
- |
- |
serine/threonine-protein kinase bri1, putative [Ricinus communis] |
| 7 |
Hb_152439_010 |
0.3002406642 |
- |
- |
PREDICTED: uncharacterized protein LOC105796032 [Gossypium raimondii] |
| 8 |
Hb_002188_020 |
0.3091878815 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 9 |
Hb_178613_010 |
0.3103924206 |
- |
- |
PREDICTED: uncharacterized protein LOC105796032 [Gossypium raimondii] |
| 10 |
Hb_128287_010 |
0.3190330215 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 11 |
Hb_000380_200 |
0.3193678813 |
- |
- |
- |
| 12 |
Hb_108404_010 |
0.3199551439 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 13 |
Hb_000204_180 |
0.3204836174 |
- |
- |
PREDICTED: uncharacterized protein LOC104094989 [Nicotiana tomentosiformis] |
| 14 |
Hb_000227_110 |
0.3241276109 |
- |
- |
hypothetical protein CICLE_v10010500mg, partial [Citrus clementina] |
| 15 |
Hb_002141_010 |
0.3262902631 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 16 |
Hb_002673_050 |
0.326807641 |
- |
- |
- |
| 17 |
Hb_091369_010 |
0.3272309982 |
- |
- |
- |
| 18 |
Hb_008473_010 |
0.3274397943 |
- |
- |
PREDICTED: laccase-14-like [Jatropha curcas] |
| 19 |
Hb_000076_170 |
0.3296399482 |
- |
- |
hypothetical protein JCGZ_00832 [Jatropha curcas] |
| 20 |
Hb_000944_010 |
0.3298664705 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |