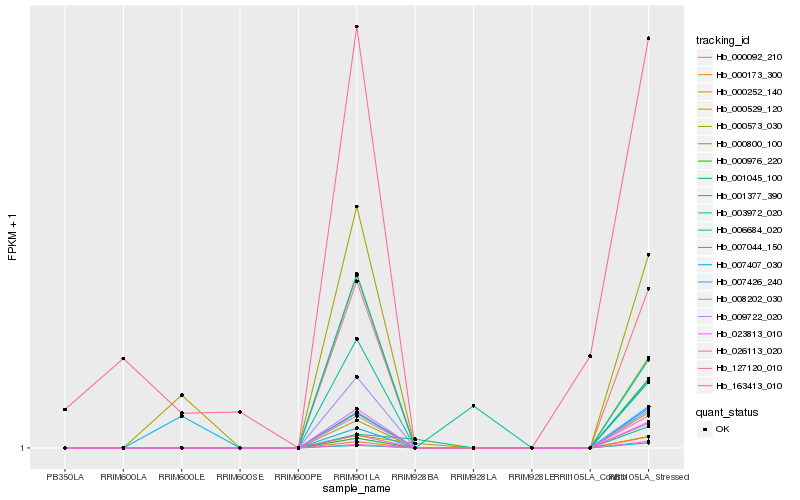
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007426_240 |
0.0 |
- |
- |
- |
| 2 |
Hb_000529_120 |
0.0220326102 |
- |
- |
PREDICTED: uncharacterized protein LOC105641745 [Jatropha curcas] |
| 3 |
Hb_000092_210 |
0.0433488643 |
- |
- |
- |
| 4 |
Hb_023813_010 |
0.0749126792 |
- |
- |
PREDICTED: uncharacterized protein LOC105162193 [Sesamum indicum] |
| 5 |
Hb_009722_020 |
0.0895987177 |
- |
- |
- |
| 6 |
Hb_000173_300 |
0.0917045724 |
- |
- |
PREDICTED: uncharacterized protein LOC105631894 [Jatropha curcas] |
| 7 |
Hb_026113_020 |
0.1024531416 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 8 |
Hb_006684_020 |
0.1055977165 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 9 |
Hb_000976_220 |
0.1164826298 |
- |
- |
PREDICTED: protein ALTERED XYLOGLUCAN 4-like [Jatropha curcas] |
| 10 |
Hb_127120_010 |
0.1165358019 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 11 |
Hb_007407_030 |
0.116732279 |
- |
- |
hypothetical protein JCGZ_23510 [Jatropha curcas] |
| 12 |
Hb_008202_030 |
0.171099681 |
- |
- |
hypothetical protein B456_002G115600 [Gossypium raimondii] |
| 13 |
Hb_000800_100 |
0.1855719369 |
- |
- |
hypothetical protein JCGZ_01543 [Jatropha curcas] |
| 14 |
Hb_000573_030 |
0.235816498 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF181 [Jatropha curcas] |
| 15 |
Hb_000252_140 |
0.2952699538 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 16 |
Hb_003972_020 |
0.3087374885 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001045_100 |
0.3247028699 |
- |
- |
PREDICTED: uncharacterized protein LOC102611758 [Citrus sinensis] |
| 18 |
Hb_163413_010 |
0.3260040188 |
- |
- |
PREDICTED: uncharacterized protein LOC102604542 [Solanum tuberosum] |
| 19 |
Hb_007044_150 |
0.3317810086 |
- |
- |
PREDICTED: DNA-directed RNA polymerases II and V subunit 8A-like [Jatropha curcas] |
| 20 |
Hb_001377_390 |
0.3329883781 |
- |
- |
hypothetical protein POPTR_0002s17390g [Populus trichocarpa] |