| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
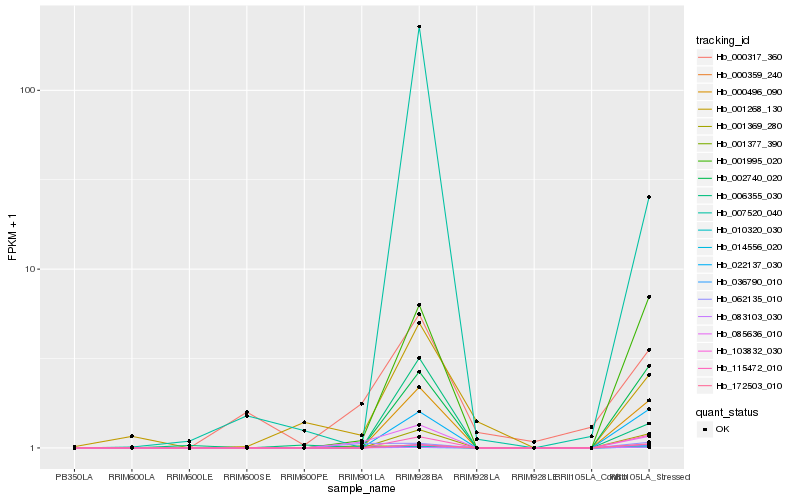
Hb_085636_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636643 [Jatropha curcas] |
| 2 |
Hb_010320_030 |
0.0695871845 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 3 |
Hb_000359_240 |
0.2450231258 |
- |
- |
PREDICTED: uncharacterized protein LOC105646039 [Jatropha curcas] |
| 4 |
Hb_062135_010 |
0.2480891025 |
- |
- |
hypothetical protein POPTR_0011s036602g, partial [Populus trichocarpa] |
| 5 |
Hb_000317_360 |
0.2727126328 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g08460 [Jatropha curcas] |
| 6 |
Hb_000496_090 |
0.2787400569 |
- |
- |
PREDICTED: inhibitor of trypsin and hageman factor-like [Gossypium raimondii] |
| 7 |
Hb_001369_280 |
0.2825570513 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g64320, mitochondrial-like isoform X3 [Solanum tuberosum] |
| 8 |
Hb_001995_020 |
0.2871001596 |
- |
- |
PREDICTED: trans-resveratrol di-O-methyltransferase-like [Populus euphratica] |
| 9 |
Hb_036790_010 |
0.2896619098 |
- |
- |
PREDICTED: GDSL esterase/lipase 1-like [Jatropha curcas] |
| 10 |
Hb_022137_030 |
0.3144383549 |
- |
- |
PREDICTED: RING-H2 finger protein ATL66 [Jatropha curcas] |
| 11 |
Hb_002740_020 |
0.3188930608 |
- |
- |
- |
| 12 |
Hb_115472_010 |
0.3214468191 |
- |
- |
- |
| 13 |
Hb_001268_130 |
0.3224003089 |
- |
- |
hypothetical protein JCGZ_26837 [Jatropha curcas] |
| 14 |
Hb_172503_010 |
0.3296853118 |
- |
- |
PREDICTED: uncharacterized protein LOC105647154 [Jatropha curcas] |
| 15 |
Hb_103832_030 |
0.3301332563 |
- |
- |
PREDICTED: uncharacterized protein LOC105795045 [Gossypium raimondii] |
| 16 |
Hb_083103_030 |
0.3323044658 |
- |
- |
putative retrotransposon protein [Malus domestica] |
| 17 |
Hb_006355_030 |
0.3337381432 |
transcription factor |
TF Family: Orphans |
sensory transduction histidine kinase bacterial, putative [Ricinus communis] |
| 18 |
Hb_014556_020 |
0.3391095407 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 19 |
Hb_001377_390 |
0.3488528152 |
- |
- |
hypothetical protein POPTR_0002s17390g [Populus trichocarpa] |
| 20 |
Hb_007520_040 |
0.3533559151 |
- |
- |
PREDICTED: carbonic anhydrase 2-like [Jatropha curcas] |