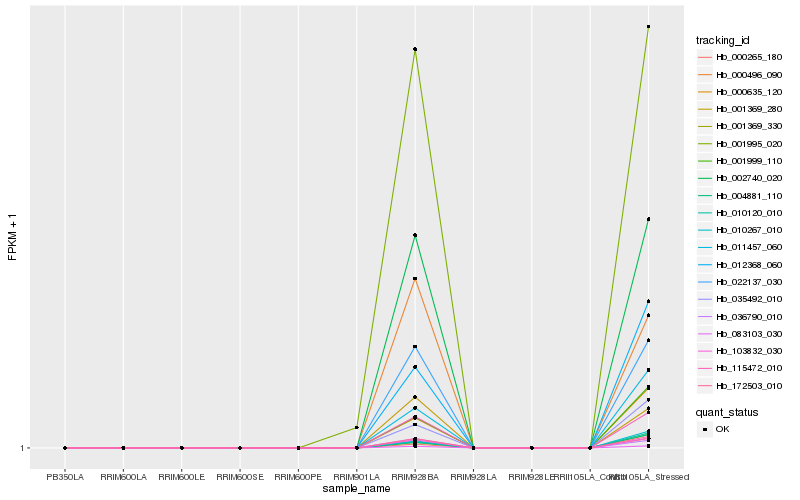
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001369_280 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g64320, mitochondrial-like isoform X3 [Solanum tuberosum] |
| 2 |
Hb_000496_090 |
0.01251578 |
- |
- |
PREDICTED: inhibitor of trypsin and hageman factor-like [Gossypium raimondii] |
| 3 |
Hb_036790_010 |
0.0202223219 |
- |
- |
PREDICTED: GDSL esterase/lipase 1-like [Jatropha curcas] |
| 4 |
Hb_022137_030 |
0.0757041602 |
- |
- |
PREDICTED: RING-H2 finger protein ATL66 [Jatropha curcas] |
| 5 |
Hb_002740_020 |
0.0843125527 |
- |
- |
- |
| 6 |
Hb_115472_010 |
0.0891243768 |
- |
- |
- |
| 7 |
Hb_172503_010 |
0.1041212961 |
- |
- |
PREDICTED: uncharacterized protein LOC105647154 [Jatropha curcas] |
| 8 |
Hb_103832_030 |
0.1049159911 |
- |
- |
PREDICTED: uncharacterized protein LOC105795045 [Gossypium raimondii] |
| 9 |
Hb_001995_020 |
0.1080087817 |
- |
- |
PREDICTED: trans-resveratrol di-O-methyltransferase-like [Populus euphratica] |
| 10 |
Hb_083103_030 |
0.1087404545 |
- |
- |
putative retrotransposon protein [Malus domestica] |
| 11 |
Hb_012368_060 |
0.2154349371 |
- |
- |
- |
| 12 |
Hb_011457_060 |
0.2158425411 |
- |
- |
PREDICTED: uncharacterized protein LOC105649957 [Jatropha curcas] |
| 13 |
Hb_001999_110 |
0.2168413959 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Populus euphratica] |
| 14 |
Hb_000635_120 |
0.2169555473 |
- |
- |
- |
| 15 |
Hb_035492_010 |
0.2204503486 |
- |
- |
PREDICTED: uncharacterized protein LOC105796032 [Gossypium raimondii] |
| 16 |
Hb_010267_010 |
0.2410725879 |
- |
- |
- |
| 17 |
Hb_010120_010 |
0.2419417877 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 18 |
Hb_001369_330 |
0.2425890411 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_004881_110 |
0.2426606187 |
- |
- |
- |
| 20 |
Hb_000265_180 |
0.244434731 |
- |
- |
Alpha/beta-Hydrolases superfamily protein, putative [Theobroma cacao] |