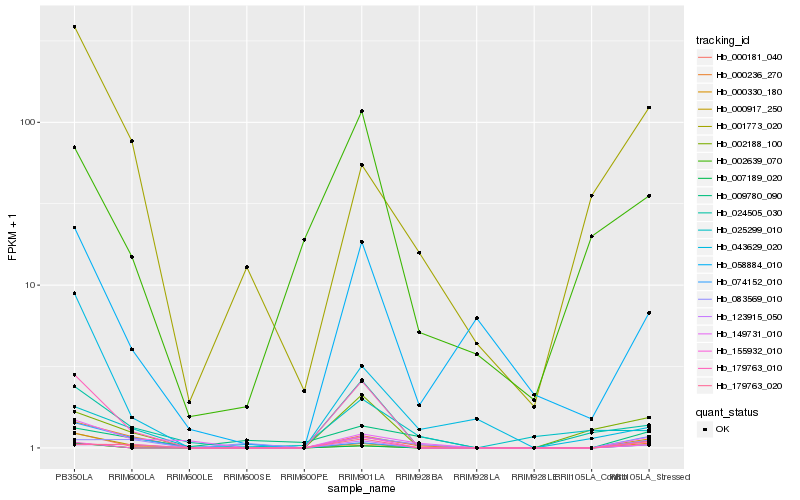
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000181_040 |
0.0 |
transcription factor |
TF Family: NAC |
PREDICTED: protein CUP-SHAPED COTYLEDON 2 [Jatropha curcas] |
| 2 |
Hb_149731_010 |
0.2690444762 |
- |
- |
- |
| 3 |
Hb_007189_020 |
0.2739111926 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 4 |
Hb_000236_270 |
0.2748609267 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 5 |
Hb_058884_010 |
0.3067740047 |
- |
- |
PREDICTED: barwin-like [Jatropha curcas] |
| 6 |
Hb_000917_250 |
0.3201688317 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 130-like [Populus euphratica] |
| 7 |
Hb_155932_010 |
0.3232614551 |
- |
- |
PREDICTED: uncharacterized protein LOC105786773 [Gossypium raimondii] |
| 8 |
Hb_009780_090 |
0.3277953409 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g66345, mitochondrial [Jatropha curcas] |
| 9 |
Hb_024505_030 |
0.3279179375 |
- |
- |
PREDICTED: aspartate aminotransferase, chloroplastic [Jatropha curcas] |
| 10 |
Hb_179763_010 |
0.3281300729 |
- |
- |
- |
| 11 |
Hb_000330_180 |
0.341817241 |
- |
- |
- |
| 12 |
Hb_001773_020 |
0.3426100401 |
- |
- |
hypothetical protein POPTR_0016s00980g [Populus trichocarpa] |
| 13 |
Hb_043629_020 |
0.3467354299 |
- |
- |
hypothetical protein POPTR_0009s13360g [Populus trichocarpa] |
| 14 |
Hb_002188_100 |
0.3474269472 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_074152_010 |
0.3487666096 |
- |
- |
- |
| 16 |
Hb_002639_070 |
0.3491438147 |
- |
- |
PREDICTED: uncharacterized protein LOC105635265 [Jatropha curcas] |
| 17 |
Hb_083569_010 |
0.3514791619 |
- |
- |
- |
| 18 |
Hb_179763_020 |
0.3517978165 |
- |
- |
- |
| 19 |
Hb_025299_010 |
0.3529347239 |
- |
- |
hypothetical protein JCGZ_01062 [Jatropha curcas] |
| 20 |
Hb_123915_050 |
0.3539261365 |
- |
- |
hypothetical protein B456_002G103400 [Gossypium raimondii] |