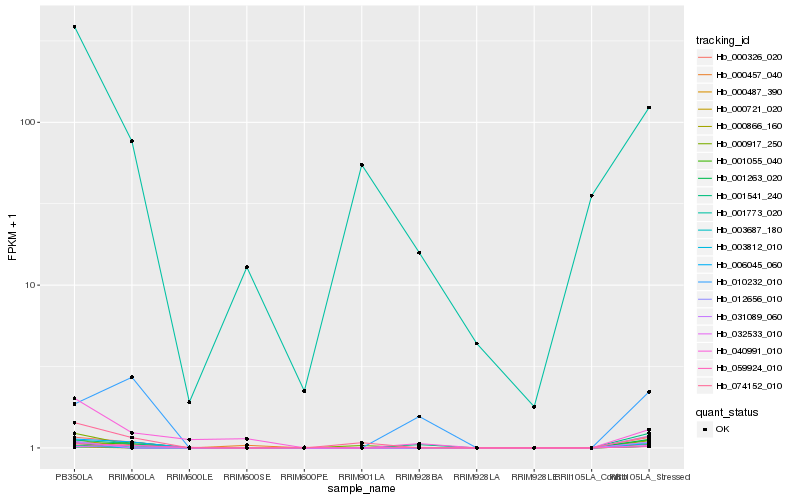
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000326_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_000721_020 |
0.1190657473 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 3 |
Hb_006045_060 |
0.1196671414 |
- |
- |
PREDICTED: uncharacterized protein LOC104222982, partial [Nicotiana sylvestris] |
| 4 |
Hb_032533_010 |
0.2156117239 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 5 |
Hb_003687_180 |
0.2253584587 |
- |
- |
hypothetical protein JCGZ_02335 [Jatropha curcas] |
| 6 |
Hb_074152_010 |
0.2679886609 |
- |
- |
- |
| 7 |
Hb_001541_240 |
0.2724225893 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000917_250 |
0.2781390757 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 130-like [Populus euphratica] |
| 9 |
Hb_000866_160 |
0.3380414757 |
- |
- |
PREDICTED: elicitor-responsive protein 3 [Populus euphratica] |
| 10 |
Hb_031089_060 |
0.3380822323 |
- |
- |
- |
| 11 |
Hb_000457_040 |
0.3389768797 |
- |
- |
- |
| 12 |
Hb_010232_010 |
0.3410538976 |
- |
- |
- |
| 13 |
Hb_000487_390 |
0.3575300456 |
- |
- |
PREDICTED: putative pumilio homolog 21 [Jatropha curcas] |
| 14 |
Hb_001263_020 |
0.3590390809 |
- |
- |
- |
| 15 |
Hb_012656_010 |
0.3594222946 |
- |
- |
- |
| 16 |
Hb_003812_010 |
0.3626190024 |
- |
- |
- |
| 17 |
Hb_059924_010 |
0.3678674128 |
- |
- |
- |
| 18 |
Hb_001773_020 |
0.3720348529 |
- |
- |
hypothetical protein POPTR_0016s00980g [Populus trichocarpa] |
| 19 |
Hb_040991_010 |
0.3739089226 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X2 [Jatropha curcas] |
| 20 |
Hb_001055_040 |
0.3821053596 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |