| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
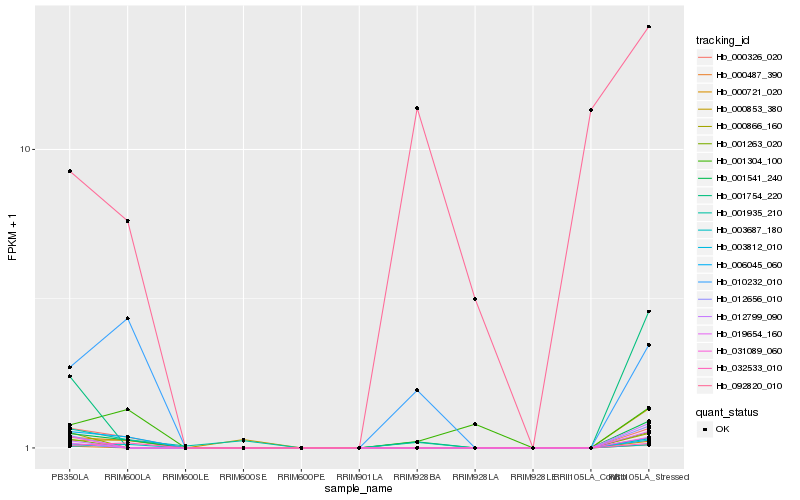
Hb_001541_240 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000326_020 |
0.2724225893 |
- |
- |
- |
| 3 |
Hb_010232_010 |
0.2726190615 |
- |
- |
- |
| 4 |
Hb_032533_010 |
0.3387100819 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 5 |
Hb_000721_020 |
0.3414607576 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 6 |
Hb_003687_180 |
0.3421851478 |
- |
- |
hypothetical protein JCGZ_02335 [Jatropha curcas] |
| 7 |
Hb_006045_060 |
0.3432007221 |
- |
- |
PREDICTED: uncharacterized protein LOC104222982, partial [Nicotiana sylvestris] |
| 8 |
Hb_003812_010 |
0.3641087873 |
- |
- |
- |
| 9 |
Hb_012656_010 |
0.3642823416 |
- |
- |
- |
| 10 |
Hb_001263_020 |
0.3643194922 |
- |
- |
- |
| 11 |
Hb_000487_390 |
0.3645035538 |
- |
- |
PREDICTED: putative pumilio homolog 21 [Jatropha curcas] |
| 12 |
Hb_001754_220 |
0.367082209 |
- |
- |
- |
| 13 |
Hb_001304_100 |
0.3763366279 |
- |
- |
- |
| 14 |
Hb_000853_380 |
0.3775690819 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_019654_160 |
0.3796494944 |
- |
- |
Zinc finger protein GIS2 [Medicago truncatula] |
| 16 |
Hb_012799_090 |
0.3800333536 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g29230-like [Jatropha curcas] |
| 17 |
Hb_000866_160 |
0.3902924553 |
- |
- |
PREDICTED: elicitor-responsive protein 3 [Populus euphratica] |
| 18 |
Hb_031089_060 |
0.3908201381 |
- |
- |
- |
| 19 |
Hb_001935_210 |
0.39553553 |
- |
- |
- |
| 20 |
Hb_092820_010 |
0.3957459345 |
- |
- |
hypothetical protein CISIN_1g033834mg [Citrus sinensis] |