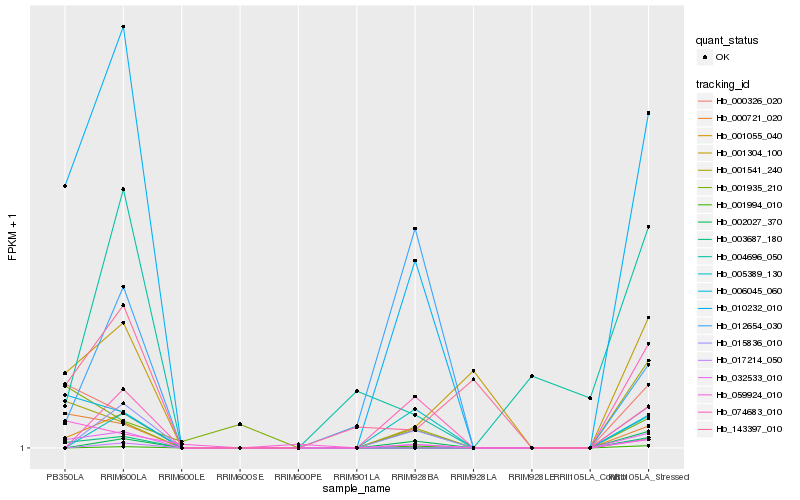
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010232_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_032533_010 |
0.2640887113 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 3 |
Hb_003687_180 |
0.2649967326 |
- |
- |
hypothetical protein JCGZ_02335 [Jatropha curcas] |
| 4 |
Hb_001541_240 |
0.2726190615 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001055_040 |
0.3100587686 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 6 |
Hb_000721_020 |
0.3176874472 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 7 |
Hb_006045_060 |
0.3227382819 |
- |
- |
PREDICTED: uncharacterized protein LOC104222982, partial [Nicotiana sylvestris] |
| 8 |
Hb_001304_100 |
0.3303639646 |
- |
- |
- |
| 9 |
Hb_015836_010 |
0.330432303 |
- |
- |
- |
| 10 |
Hb_000326_020 |
0.3410538976 |
- |
- |
- |
| 11 |
Hb_017214_050 |
0.3694389525 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 12 |
Hb_002027_370 |
0.3701134068 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_059924_010 |
0.3760297204 |
- |
- |
- |
| 14 |
Hb_074683_010 |
0.376563335 |
- |
- |
- |
| 15 |
Hb_005389_130 |
0.3811609676 |
- |
- |
hypothetical protein CICLE_v10009852mg [Citrus clementina] |
| 16 |
Hb_012654_030 |
0.3856870176 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001935_210 |
0.3947933424 |
- |
- |
- |
| 18 |
Hb_001994_010 |
0.3983191612 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 19 |
Hb_004696_050 |
0.3999432845 |
- |
- |
- |
| 20 |
Hb_143397_010 |
0.4051361178 |
- |
- |
PREDICTED: uncharacterized protein LOC105637052 [Jatropha curcas] |