| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
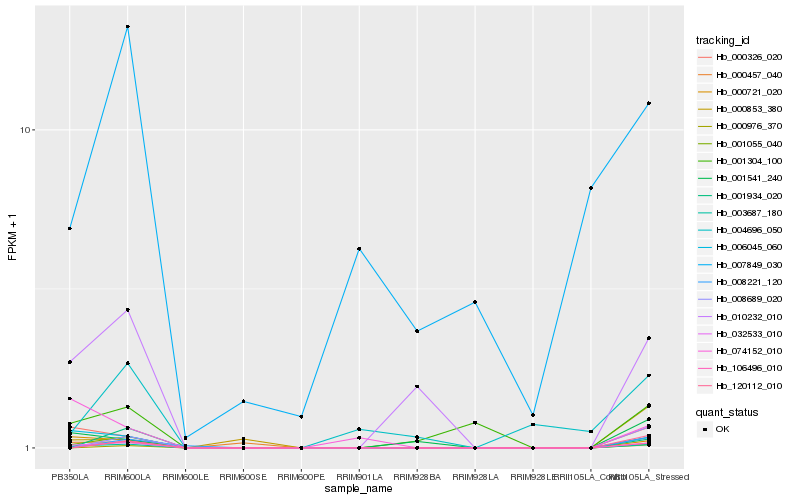
Hb_003687_180 |
0.0 |
- |
- |
hypothetical protein JCGZ_02335 [Jatropha curcas] |
| 2 |
Hb_032533_010 |
0.0114297289 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 3 |
Hb_001055_040 |
0.214572038 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 4 |
Hb_000721_020 |
0.2146212237 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 5 |
Hb_006045_060 |
0.2234937809 |
- |
- |
PREDICTED: uncharacterized protein LOC104222982, partial [Nicotiana sylvestris] |
| 6 |
Hb_000326_020 |
0.2253584587 |
- |
- |
- |
| 7 |
Hb_010232_010 |
0.2649967326 |
- |
- |
- |
| 8 |
Hb_106496_010 |
0.329811079 |
- |
- |
PREDICTED: uncharacterized protein LOC105796032 [Gossypium raimondii] |
| 9 |
Hb_000457_040 |
0.3413469679 |
- |
- |
- |
| 10 |
Hb_001541_240 |
0.3421851478 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001304_100 |
0.3460761923 |
- |
- |
- |
| 12 |
Hb_000976_370 |
0.3546277409 |
- |
- |
beta-glucosidase, putative [Ricinus communis] |
| 13 |
Hb_120112_010 |
0.3561573889 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 14 |
Hb_001934_020 |
0.3571296207 |
- |
- |
PREDICTED: uncharacterized protein LOC105178413 [Sesamum indicum] |
| 15 |
Hb_008689_020 |
0.3574297547 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_074152_010 |
0.3859862742 |
- |
- |
- |
| 17 |
Hb_008221_120 |
0.3939130142 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g63170-like [Jatropha curcas] |
| 18 |
Hb_004696_050 |
0.4015767536 |
- |
- |
- |
| 19 |
Hb_000853_380 |
0.4030773128 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_007849_030 |
0.4079772644 |
- |
- |
l-allo-threonine aldolase, putative [Ricinus communis] |