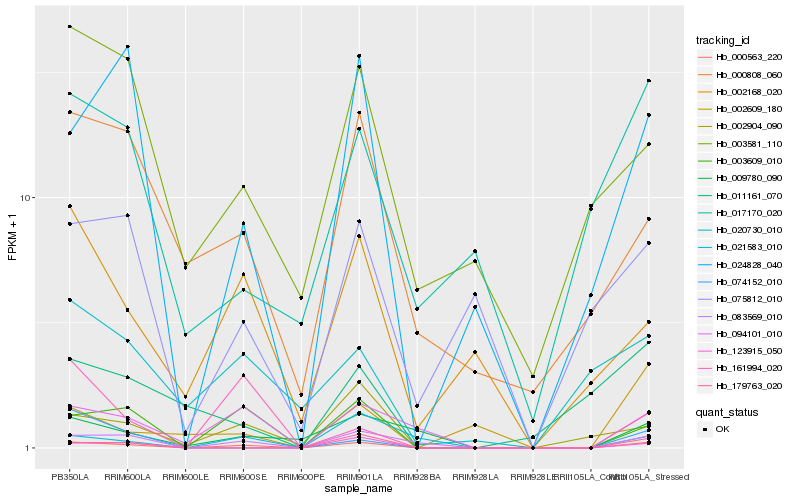
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000563_220 |
0.0 |
- |
- |
hypothetical protein VITISV_004111 [Vitis vinifera] |
| 2 |
Hb_003609_010 |
0.2167054761 |
- |
- |
- |
| 3 |
Hb_083569_010 |
0.222215683 |
- |
- |
- |
| 4 |
Hb_094101_010 |
0.2612528285 |
- |
- |
hypothetical protein JCGZ_18414 [Jatropha curcas] |
| 5 |
Hb_024828_040 |
0.2783603512 |
- |
- |
- |
| 6 |
Hb_002904_090 |
0.2825184537 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g71420-like [Vitis vinifera] |
| 7 |
Hb_179763_020 |
0.2895045857 |
- |
- |
- |
| 8 |
Hb_123915_050 |
0.2980859918 |
- |
- |
hypothetical protein B456_002G103400 [Gossypium raimondii] |
| 9 |
Hb_002168_020 |
0.3164887645 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 10 |
Hb_009780_090 |
0.3201896486 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g66345, mitochondrial [Jatropha curcas] |
| 11 |
Hb_161994_020 |
0.3204730694 |
- |
- |
- |
| 12 |
Hb_000808_060 |
0.3225396754 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 16-like [Jatropha curcas] |
| 13 |
Hb_020730_010 |
0.3275357332 |
- |
- |
hypothetical protein CICLE_v10010243mg, partial [Citrus clementina] |
| 14 |
Hb_021583_010 |
0.3295959229 |
- |
- |
- |
| 15 |
Hb_075812_010 |
0.3381377438 |
- |
- |
hypothetical protein POPTR_0005s04330g [Populus trichocarpa] |
| 16 |
Hb_003581_110 |
0.3381452179 |
- |
- |
PREDICTED: uncharacterized protein LOC105649778 isoform X2 [Jatropha curcas] |
| 17 |
Hb_002609_180 |
0.3397580442 |
- |
- |
- |
| 18 |
Hb_017170_020 |
0.3410400775 |
- |
- |
PREDICTED: OPA3-like protein isoform X2 [Jatropha curcas] |
| 19 |
Hb_074152_010 |
0.3433471526 |
- |
- |
- |
| 20 |
Hb_011161_070 |
0.3452877595 |
- |
- |
PREDICTED: kynurenine--oxoglutarate transaminase 1-like isoform X1 [Populus euphratica] |