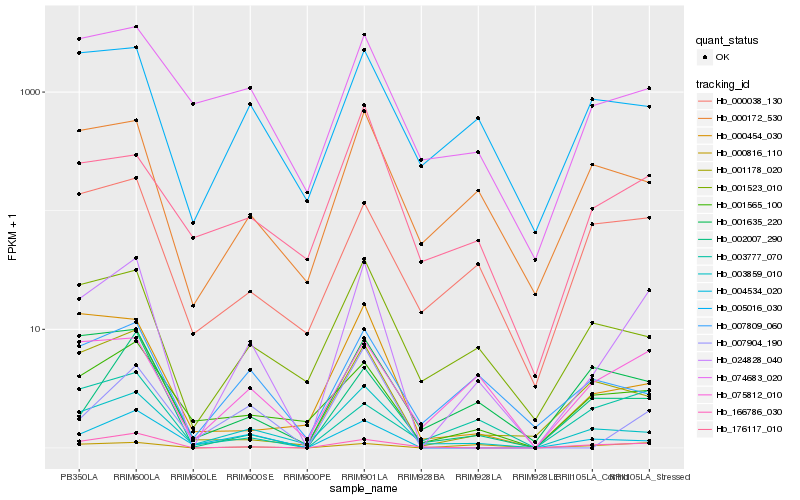
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_024828_040 |
0.0 |
- |
- |
- |
| 2 |
Hb_166786_030 |
0.2138651541 |
- |
- |
PREDICTED: uncharacterized protein LOC105775425 [Gossypium raimondii] |
| 3 |
Hb_003859_010 |
0.2197507293 |
- |
- |
PREDICTED: uncharacterized protein LOC104647507 [Solanum lycopersicum] |
| 4 |
Hb_075812_010 |
0.2282144843 |
- |
- |
hypothetical protein POPTR_0005s04330g [Populus trichocarpa] |
| 5 |
Hb_001565_100 |
0.242868035 |
- |
- |
unnamed protein product [Coffea canephora] |
| 6 |
Hb_004534_020 |
0.2486544984 |
- |
- |
PREDICTED: cysteine proteinase inhibitor B [Jatropha curcas] |
| 7 |
Hb_000816_110 |
0.2497745275 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750-like [Jatropha curcas] |
| 8 |
Hb_001523_010 |
0.2521458368 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_25496 [Jatropha curcas] |
| 9 |
Hb_001635_220 |
0.2555067752 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH87 [Jatropha curcas] |
| 10 |
Hb_001178_020 |
0.2592202643 |
transcription factor |
TF Family: GRF |
PREDICTED: growth-regulating factor 3 [Jatropha curcas] |
| 11 |
Hb_002007_290 |
0.2615078587 |
- |
- |
- |
| 12 |
Hb_000172_530 |
0.2615329317 |
- |
- |
PREDICTED: UBP1-associated protein 2B-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_000038_130 |
0.2640445445 |
- |
- |
- |
| 14 |
Hb_005016_030 |
0.2647800228 |
- |
- |
hypothetical protein JCGZ_16549 [Jatropha curcas] |
| 15 |
Hb_003777_070 |
0.2656016714 |
- |
- |
- |
| 16 |
Hb_007904_190 |
0.2662300905 |
- |
- |
- |
| 17 |
Hb_074683_020 |
0.2672202705 |
- |
- |
PREDICTED: 60S ribosomal protein L27-3 [Jatropha curcas] |
| 18 |
Hb_000454_030 |
0.2689808392 |
- |
- |
- |
| 19 |
Hb_176117_010 |
0.2689947301 |
- |
- |
Cold-inducible RNA-binding protein [Morus notabilis] |
| 20 |
Hb_007809_060 |
0.2691374825 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH3 [Jatropha curcas] |