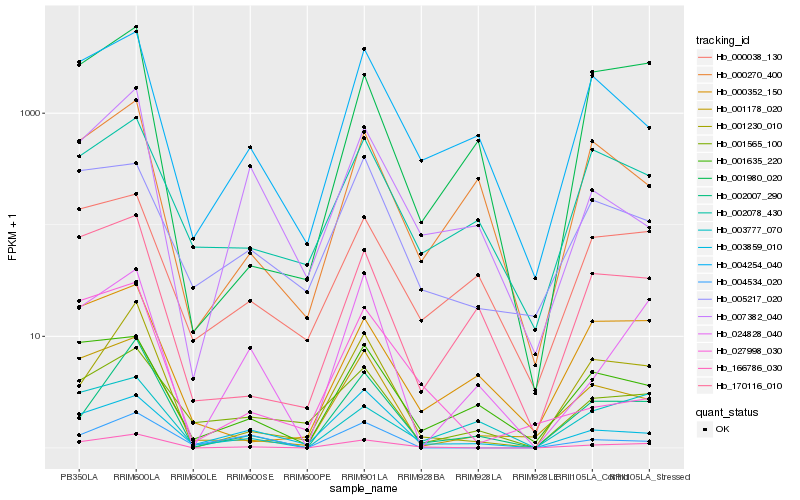
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_166786_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105775425 [Gossypium raimondii] |
| 2 |
Hb_001230_010 |
0.1828041226 |
- |
- |
- |
| 3 |
Hb_001178_020 |
0.1962674313 |
transcription factor |
TF Family: GRF |
PREDICTED: growth-regulating factor 3 [Jatropha curcas] |
| 4 |
Hb_002007_290 |
0.2026868051 |
- |
- |
- |
| 5 |
Hb_004254_040 |
0.2107571596 |
- |
- |
PREDICTED: uncharacterized protein LOC105650678 [Jatropha curcas] |
| 6 |
Hb_024828_040 |
0.2138651541 |
- |
- |
- |
| 7 |
Hb_001635_220 |
0.2178513302 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH87 [Jatropha curcas] |
| 8 |
Hb_170116_010 |
0.219037699 |
- |
- |
Geranyl diphosphate synthase 1 isoform 2 [Theobroma cacao] |
| 9 |
Hb_001980_020 |
0.2219749528 |
- |
- |
- |
| 10 |
Hb_003859_010 |
0.2241511236 |
- |
- |
PREDICTED: uncharacterized protein LOC104647507 [Solanum lycopersicum] |
| 11 |
Hb_001565_100 |
0.2271156038 |
- |
- |
unnamed protein product [Coffea canephora] |
| 12 |
Hb_000038_130 |
0.2309370985 |
- |
- |
- |
| 13 |
Hb_005217_020 |
0.2316609543 |
- |
- |
PREDICTED: ribosomal RNA processing protein 36 homolog isoform X2 [Jatropha curcas] |
| 14 |
Hb_000352_150 |
0.2344148331 |
- |
- |
hypothetical protein JCGZ_11641 [Jatropha curcas] |
| 15 |
Hb_004534_020 |
0.2347748854 |
- |
- |
PREDICTED: cysteine proteinase inhibitor B [Jatropha curcas] |
| 16 |
Hb_002078_430 |
0.2349028604 |
- |
- |
hypothetical protein PRUPE_ppa014280mg [Prunus persica] |
| 17 |
Hb_007382_040 |
0.2356493267 |
- |
- |
uncharacterized LOC105644857 [Jatropha curcas] |
| 18 |
Hb_027998_030 |
0.2388683723 |
- |
- |
laccase, putative [Ricinus communis] |
| 19 |
Hb_000270_400 |
0.2425855996 |
- |
- |
lipase, putative [Ricinus communis] |
| 20 |
Hb_003777_070 |
0.2443387925 |
- |
- |
- |