| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
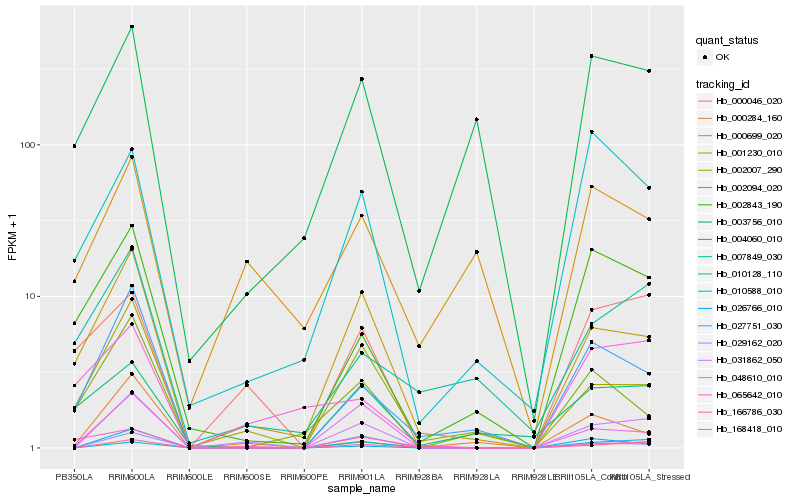
Hb_031862_050 |
0.0 |
- |
- |
Cell division cycle protein, putative [Ricinus communis] |
| 2 |
Hb_001230_010 |
0.2061595935 |
- |
- |
- |
| 3 |
Hb_002007_290 |
0.2137325454 |
- |
- |
- |
| 4 |
Hb_168418_010 |
0.2203747792 |
- |
- |
PREDICTED: uncharacterized protein LOC104234330 [Nicotiana sylvestris] |
| 5 |
Hb_000046_020 |
0.2310330427 |
- |
- |
- |
| 6 |
Hb_029162_020 |
0.2337245964 |
- |
- |
- |
| 7 |
Hb_048610_010 |
0.2403790711 |
- |
- |
- |
| 8 |
Hb_027751_030 |
0.251356459 |
- |
- |
- |
| 9 |
Hb_010588_010 |
0.2661703895 |
- |
- |
- |
| 10 |
Hb_026766_010 |
0.2677673438 |
- |
- |
- |
| 11 |
Hb_002843_190 |
0.2745887275 |
transcription factor |
TF Family: GRF |
PREDICTED: growth-regulating factor 7 isoform X1 [Jatropha curcas] |
| 12 |
Hb_010128_110 |
0.2866756632 |
- |
- |
PREDICTED: uncharacterized protein LOC105632071 [Jatropha curcas] |
| 13 |
Hb_003756_010 |
0.2875476057 |
- |
- |
PREDICTED: uncharacterized protein LOC105642108 [Jatropha curcas] |
| 14 |
Hb_065642_010 |
0.2886792278 |
- |
- |
- |
| 15 |
Hb_007849_030 |
0.293852773 |
- |
- |
l-allo-threonine aldolase, putative [Ricinus communis] |
| 16 |
Hb_004060_010 |
0.2954308439 |
- |
- |
- |
| 17 |
Hb_000699_020 |
0.2957534667 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxylase kinase 2-like [Jatropha curcas] |
| 18 |
Hb_002094_020 |
0.2961487746 |
- |
- |
PREDICTED: alpha-copaene synthase-like [Jatropha curcas] |
| 19 |
Hb_000284_160 |
0.2977342659 |
- |
- |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 20 |
Hb_166786_030 |
0.3017514476 |
- |
- |
PREDICTED: uncharacterized protein LOC105775425 [Gossypium raimondii] |