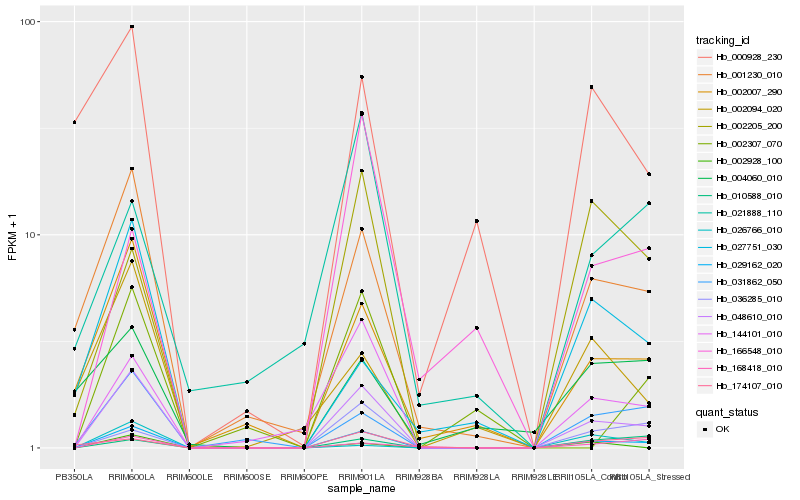
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029162_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_048610_010 |
0.0194140455 |
- |
- |
- |
| 3 |
Hb_144101_010 |
0.2212570253 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Vitis vinifera] |
| 4 |
Hb_001230_010 |
0.2212960203 |
- |
- |
- |
| 5 |
Hb_031862_050 |
0.2337245964 |
- |
- |
Cell division cycle protein, putative [Ricinus communis] |
| 6 |
Hb_002007_290 |
0.2369222892 |
- |
- |
- |
| 7 |
Hb_168418_010 |
0.2757101323 |
- |
- |
PREDICTED: uncharacterized protein LOC104234330 [Nicotiana sylvestris] |
| 8 |
Hb_036285_010 |
0.2778632379 |
- |
- |
- |
| 9 |
Hb_002205_200 |
0.284665695 |
- |
- |
PREDICTED: uncharacterized protein LOC105649174 [Jatropha curcas] |
| 10 |
Hb_010588_010 |
0.2874017398 |
- |
- |
- |
| 11 |
Hb_002928_100 |
0.2884065315 |
- |
- |
- |
| 12 |
Hb_002094_020 |
0.3028945524 |
- |
- |
PREDICTED: alpha-copaene synthase-like [Jatropha curcas] |
| 13 |
Hb_174107_010 |
0.3072804563 |
- |
- |
PREDICTED: uncharacterized protein LOC105775323 [Gossypium raimondii] |
| 14 |
Hb_002307_070 |
0.3175051478 |
- |
- |
hypothetical protein JCGZ_14841 [Jatropha curcas] |
| 15 |
Hb_027751_030 |
0.3190842049 |
- |
- |
- |
| 16 |
Hb_000928_230 |
0.3238213019 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_026766_010 |
0.327320894 |
- |
- |
- |
| 18 |
Hb_166548_010 |
0.3282388275 |
- |
- |
hypothetical protein JCGZ_09681 [Jatropha curcas] |
| 19 |
Hb_021888_110 |
0.3291796282 |
- |
- |
- |
| 20 |
Hb_004060_010 |
0.3331229463 |
- |
- |
- |