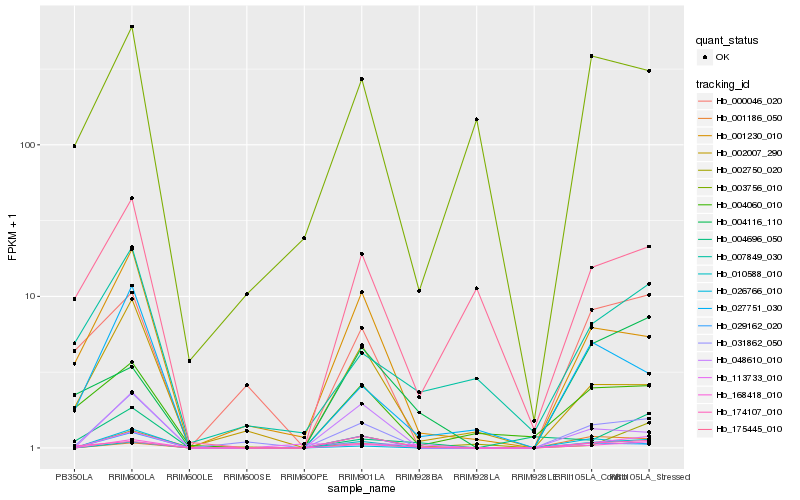
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_168418_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104234330 [Nicotiana sylvestris] |
| 2 |
Hb_031862_050 |
0.2203747792 |
- |
- |
Cell division cycle protein, putative [Ricinus communis] |
| 3 |
Hb_010588_010 |
0.2277921257 |
- |
- |
- |
| 4 |
Hb_113733_010 |
0.2351050312 |
- |
- |
hypothetical protein JCGZ_26531 [Jatropha curcas] |
| 5 |
Hb_029162_020 |
0.2757101323 |
- |
- |
- |
| 6 |
Hb_002750_020 |
0.2839894645 |
- |
- |
PREDICTED: tRNA-dihydrouridine(16/17) synthase [NAD(P)(+)]-like [Jatropha curcas] |
| 7 |
Hb_048610_010 |
0.2842077567 |
- |
- |
- |
| 8 |
Hb_001230_010 |
0.2939413286 |
- |
- |
- |
| 9 |
Hb_007849_030 |
0.2949555685 |
- |
- |
l-allo-threonine aldolase, putative [Ricinus communis] |
| 10 |
Hb_174107_010 |
0.2974791732 |
- |
- |
PREDICTED: uncharacterized protein LOC105775323 [Gossypium raimondii] |
| 11 |
Hb_004696_050 |
0.2994394843 |
- |
- |
- |
| 12 |
Hb_027751_030 |
0.3011109474 |
- |
- |
- |
| 13 |
Hb_004116_110 |
0.309764931 |
- |
- |
hypothetical protein MTR_3g064100 [Medicago truncatula] |
| 14 |
Hb_002007_290 |
0.3110112583 |
- |
- |
- |
| 15 |
Hb_175445_010 |
0.3149629244 |
- |
- |
Pectin methylesterase 31 isoform 2, partial [Theobroma cacao] |
| 16 |
Hb_004060_010 |
0.3198754175 |
- |
- |
- |
| 17 |
Hb_026766_010 |
0.3224605211 |
- |
- |
- |
| 18 |
Hb_000046_020 |
0.3232031549 |
- |
- |
- |
| 19 |
Hb_003756_010 |
0.3286794063 |
- |
- |
PREDICTED: uncharacterized protein LOC105642108 [Jatropha curcas] |
| 20 |
Hb_001186_050 |
0.3287803161 |
- |
- |
- |