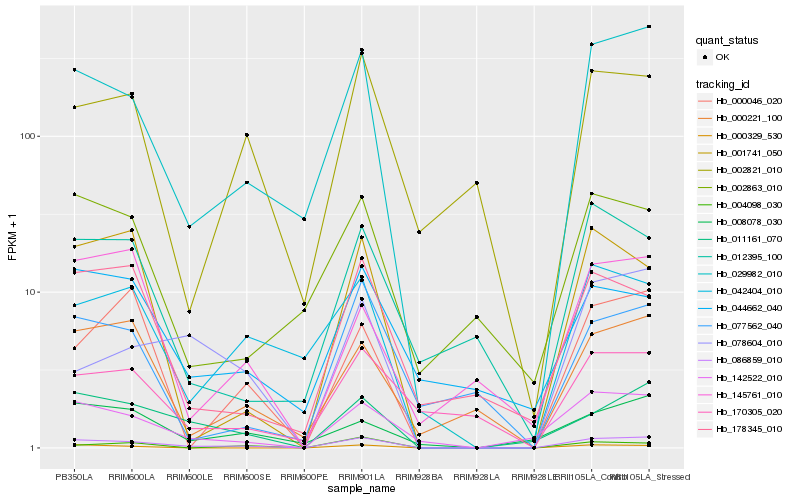
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_086859_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_029982_010 |
0.1233308021 |
- |
- |
hypothetical protein CICLE_v10006145mg [Citrus clementina] |
| 3 |
Hb_011161_070 |
0.169082827 |
- |
- |
PREDICTED: kynurenine--oxoglutarate transaminase 1-like isoform X1 [Populus euphratica] |
| 4 |
Hb_142522_010 |
0.1754679713 |
- |
- |
- |
| 5 |
Hb_042404_010 |
0.1954173926 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |
| 6 |
Hb_000221_100 |
0.2050269156 |
- |
- |
hypothetical protein POPTR_0006s07000g [Populus trichocarpa] |
| 7 |
Hb_000329_530 |
0.205716791 |
- |
- |
Uncharacterized protein TCM_038736 [Theobroma cacao] |
| 8 |
Hb_001741_050 |
0.2105596766 |
- |
- |
- |
| 9 |
Hb_000046_020 |
0.2196118547 |
- |
- |
- |
| 10 |
Hb_178345_010 |
0.220009606 |
- |
- |
hypothetical protein PHAVU_004G084200g [Phaseolus vulgaris] |
| 11 |
Hb_078604_010 |
0.2212696434 |
- |
- |
- |
| 12 |
Hb_008078_030 |
0.2219844471 |
- |
- |
- |
| 13 |
Hb_004098_030 |
0.2224036796 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 14 |
Hb_002821_010 |
0.2228909797 |
- |
- |
PREDICTED: nucleolar protein 16 [Jatropha curcas] |
| 15 |
Hb_012395_100 |
0.2246644281 |
- |
- |
eukaryotic translation initiation factor 4e, putative [Ricinus communis] |
| 16 |
Hb_170305_020 |
0.2252699403 |
- |
- |
hypothetical protein JCGZ_23578 [Jatropha curcas] |
| 17 |
Hb_044662_040 |
0.226044275 |
- |
- |
- |
| 18 |
Hb_077562_040 |
0.2295440595 |
- |
- |
hypothetical protein JCGZ_24573 [Jatropha curcas] |
| 19 |
Hb_002863_010 |
0.2309462531 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RNF144A-B [Jatropha curcas] |
| 20 |
Hb_145761_010 |
0.2324662066 |
- |
- |
- |