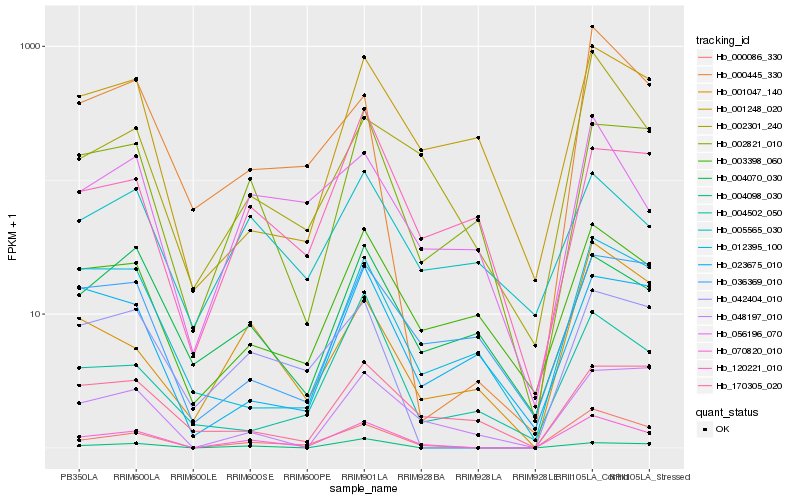
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_070820_010 |
0.0 |
- |
- |
hypothetical protein B456_008G232800 [Gossypium raimondii] |
| 2 |
Hb_000086_330 |
0.115091138 |
- |
- |
PREDICTED: glutathione gamma-glutamylcysteinyltransferase 1 [Jatropha curcas] |
| 3 |
Hb_002821_010 |
0.1929475009 |
- |
- |
PREDICTED: nucleolar protein 16 [Jatropha curcas] |
| 4 |
Hb_004098_030 |
0.1930228096 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 5 |
Hb_042404_010 |
0.2033172354 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |
| 6 |
Hb_003398_060 |
0.205755449 |
- |
- |
PREDICTED: rRNA methyltransferase 2, mitochondrial isoform X1 [Jatropha curcas] |
| 7 |
Hb_048197_010 |
0.2064767229 |
- |
- |
- |
| 8 |
Hb_002301_240 |
0.2071365187 |
- |
- |
auxin-repressed protein-like protein ARP1 [Manihot esculenta] |
| 9 |
Hb_000445_330 |
0.2205514509 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001047_140 |
0.2210832992 |
- |
- |
- |
| 11 |
Hb_001248_020 |
0.2250622743 |
- |
- |
PREDICTED: uncharacterized protein At2g23090-like [Jatropha curcas] |
| 12 |
Hb_056196_070 |
0.225992954 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA26-like [Jatropha curcas] |
| 13 |
Hb_012395_100 |
0.2274310349 |
- |
- |
eukaryotic translation initiation factor 4e, putative [Ricinus communis] |
| 14 |
Hb_036369_010 |
0.2326927051 |
- |
- |
- |
| 15 |
Hb_004070_030 |
0.2340758341 |
- |
- |
PREDICTED: phosphatidate cytidylyltransferase, mitochondrial isoform X2 [Jatropha curcas] |
| 16 |
Hb_004502_050 |
0.2363168627 |
- |
- |
hypothetical protein POPTR_1488s00200g [Populus trichocarpa] |
| 17 |
Hb_005565_030 |
0.2378809188 |
- |
- |
PREDICTED: uncharacterized protein LOC105633107 isoform X1 [Jatropha curcas] |
| 18 |
Hb_120221_010 |
0.2385058667 |
- |
- |
- |
| 19 |
Hb_170305_020 |
0.2401228304 |
- |
- |
hypothetical protein JCGZ_23578 [Jatropha curcas] |
| 20 |
Hb_023675_010 |
0.2411805784 |
- |
- |
- |