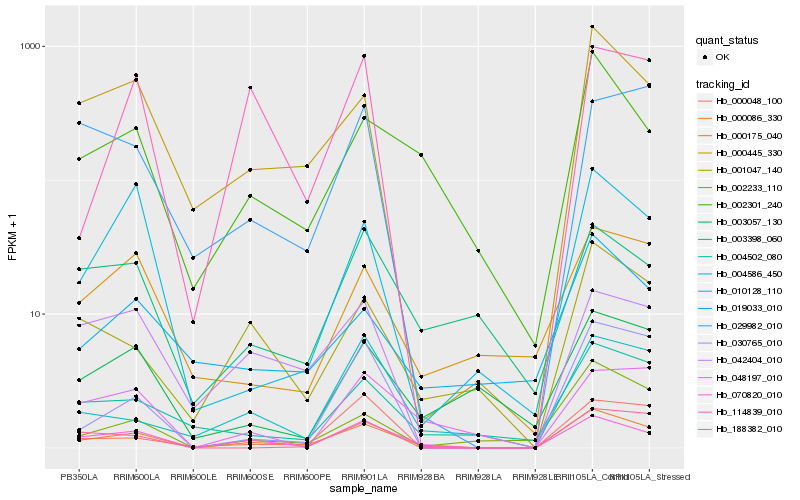
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000086_330 |
0.0 |
- |
- |
PREDICTED: glutathione gamma-glutamylcysteinyltransferase 1 [Jatropha curcas] |
| 2 |
Hb_070820_010 |
0.115091138 |
- |
- |
hypothetical protein B456_008G232800 [Gossypium raimondii] |
| 3 |
Hb_000445_330 |
0.1842758215 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002301_240 |
0.1882719861 |
- |
- |
auxin-repressed protein-like protein ARP1 [Manihot esculenta] |
| 5 |
Hb_001047_140 |
0.1945538519 |
- |
- |
- |
| 6 |
Hb_004502_080 |
0.2032071261 |
- |
- |
hypothetical protein JCGZ_23578 [Jatropha curcas] |
| 7 |
Hb_030765_010 |
0.2119707724 |
desease resistance |
Gene Name: ABC_tran |
PREDICTED: ABC transporter G family member 11-like isoform X1 [Solanum lycopersicum] |
| 8 |
Hb_042404_010 |
0.216329445 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |
| 9 |
Hb_002233_110 |
0.2175761298 |
- |
- |
PREDICTED: nuclear transcription factor Y subunit B-3-like [Jatropha curcas] |
| 10 |
Hb_004586_450 |
0.2180910154 |
- |
- |
- |
| 11 |
Hb_188382_010 |
0.2217077917 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 12 |
Hb_048197_010 |
0.2236064565 |
- |
- |
- |
| 13 |
Hb_114839_010 |
0.2291920009 |
- |
- |
- |
| 14 |
Hb_010128_110 |
0.2292751643 |
- |
- |
PREDICTED: uncharacterized protein LOC105632071 [Jatropha curcas] |
| 15 |
Hb_000048_100 |
0.2303919172 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_019033_010 |
0.2306620014 |
- |
- |
PREDICTED: uncharacterized protein LOC105643397 [Jatropha curcas] |
| 17 |
Hb_003057_130 |
0.2324542197 |
- |
- |
- |
| 18 |
Hb_000175_040 |
0.2346828633 |
- |
- |
hypothetical protein JCGZ_06119 [Jatropha curcas] |
| 19 |
Hb_003398_060 |
0.2355978572 |
- |
- |
PREDICTED: rRNA methyltransferase 2, mitochondrial isoform X1 [Jatropha curcas] |
| 20 |
Hb_029982_010 |
0.2407095176 |
- |
- |
hypothetical protein CICLE_v10006145mg [Citrus clementina] |