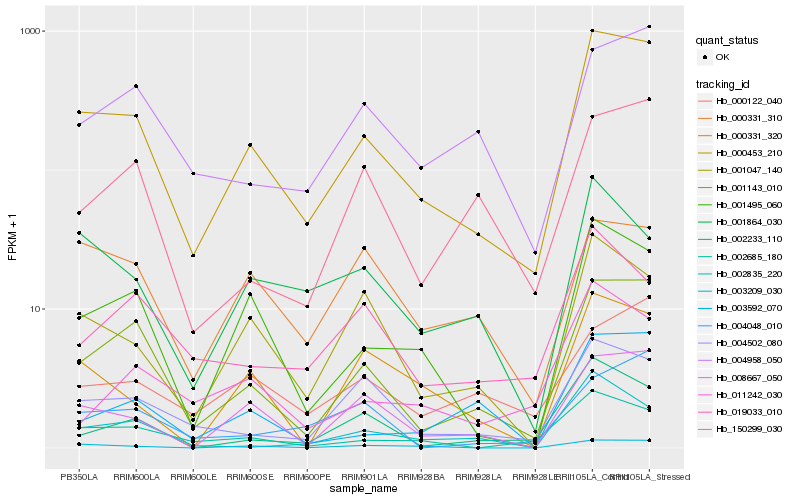
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000453_210 |
0.0 |
- |
- |
hypothetical protein PHAVU_006G019800g [Phaseolus vulgaris] |
| 2 |
Hb_001143_010 |
0.1244241929 |
- |
- |
- |
| 3 |
Hb_001495_060 |
0.1540596622 |
- |
- |
PREDICTED: probable aldo-keto reductase 1 [Jatropha curcas] |
| 4 |
Hb_002685_180 |
0.1626787343 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 5 |
Hb_008667_050 |
0.165938311 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Malus domestica] |
| 6 |
Hb_003209_030 |
0.1751539576 |
- |
- |
PREDICTED: heparanase-like protein 2 [Jatropha curcas] |
| 7 |
Hb_001047_140 |
0.1769370791 |
- |
- |
- |
| 8 |
Hb_004502_080 |
0.1797280992 |
- |
- |
hypothetical protein JCGZ_23578 [Jatropha curcas] |
| 9 |
Hb_004958_050 |
0.1979080972 |
- |
- |
hypothetical protein CICLE_v10032980mg [Citrus clementina] |
| 10 |
Hb_000331_320 |
0.1993140727 |
- |
- |
- |
| 11 |
Hb_000122_040 |
0.2008682407 |
- |
- |
PREDICTED: uncharacterized protein LOC105649224 [Jatropha curcas] |
| 12 |
Hb_002233_110 |
0.2050803837 |
- |
- |
PREDICTED: nuclear transcription factor Y subunit B-3-like [Jatropha curcas] |
| 13 |
Hb_001864_030 |
0.2084707113 |
- |
- |
thioredoxin h [Hevea brasiliensis] |
| 14 |
Hb_150299_030 |
0.2099516342 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_019033_010 |
0.2130540873 |
- |
- |
PREDICTED: uncharacterized protein LOC105643397 [Jatropha curcas] |
| 16 |
Hb_003592_070 |
0.2140779701 |
- |
- |
- |
| 17 |
Hb_000331_310 |
0.2145507088 |
- |
- |
PREDICTED: uncharacterized protein LOC105640232 [Jatropha curcas] |
| 18 |
Hb_002835_220 |
0.2153322241 |
- |
- |
PREDICTED: MATE efflux family protein 7 [Vitis vinifera] |
| 19 |
Hb_011242_030 |
0.2153594393 |
- |
- |
- |
| 20 |
Hb_004048_010 |
0.2158246275 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570-like [Citrus sinensis] |