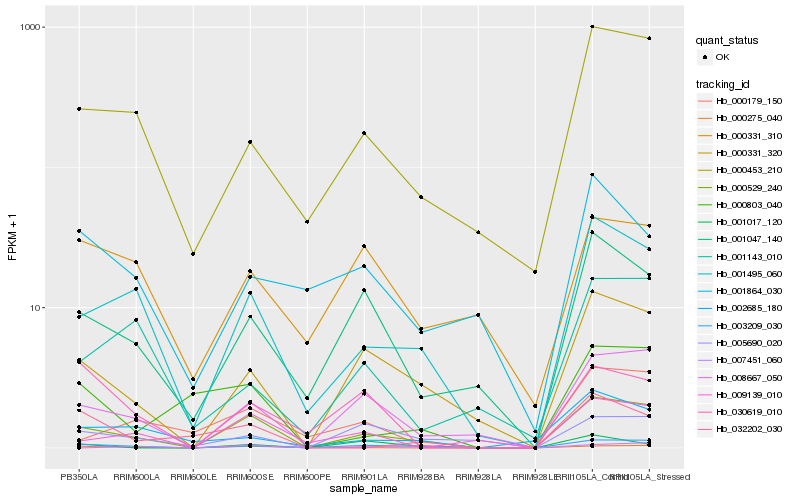
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000529_240 |
0.0 |
- |
- |
- |
| 2 |
Hb_003209_030 |
0.1756168961 |
- |
- |
PREDICTED: heparanase-like protein 2 [Jatropha curcas] |
| 3 |
Hb_001495_060 |
0.1775299213 |
- |
- |
PREDICTED: probable aldo-keto reductase 1 [Jatropha curcas] |
| 4 |
Hb_008667_050 |
0.1796893133 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Malus domestica] |
| 5 |
Hb_000331_320 |
0.1848131565 |
- |
- |
- |
| 6 |
Hb_001047_140 |
0.2244299984 |
- |
- |
- |
| 7 |
Hb_000453_210 |
0.2280229162 |
- |
- |
hypothetical protein PHAVU_006G019800g [Phaseolus vulgaris] |
| 8 |
Hb_009139_010 |
0.2446484001 |
- |
- |
putative Phospholipid-Diacylglycerol acyltransferase [Elaeis guineensis] |
| 9 |
Hb_000275_040 |
0.246994759 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 10 |
Hb_005690_020 |
0.2639140893 |
- |
- |
hypothetical protein POPTR_0006s08200g [Populus trichocarpa] |
| 11 |
Hb_000179_150 |
0.2655504247 |
- |
- |
hypothetical protein ZEAMMB73_153699 [Zea mays] |
| 12 |
Hb_000803_040 |
0.2670219454 |
- |
- |
- |
| 13 |
Hb_007451_060 |
0.2776993847 |
- |
- |
BAHD family acyltransferase, clade V, putative [Theobroma cacao] |
| 14 |
Hb_002685_180 |
0.2789481613 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 15 |
Hb_030619_010 |
0.2870435675 |
- |
- |
- |
| 16 |
Hb_000331_310 |
0.2871297566 |
- |
- |
PREDICTED: uncharacterized protein LOC105640232 [Jatropha curcas] |
| 17 |
Hb_001143_010 |
0.2906222447 |
- |
- |
- |
| 18 |
Hb_032202_030 |
0.2942694724 |
- |
- |
- |
| 19 |
Hb_001864_030 |
0.2954968405 |
- |
- |
thioredoxin h [Hevea brasiliensis] |
| 20 |
Hb_001017_120 |
0.2977837604 |
- |
- |
- |