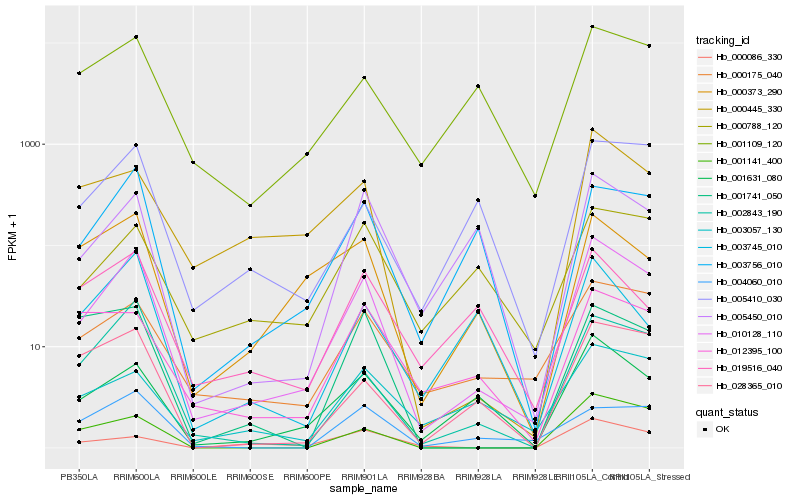
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010128_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632071 [Jatropha curcas] |
| 2 |
Hb_002843_190 |
0.1767079858 |
transcription factor |
TF Family: GRF |
PREDICTED: growth-regulating factor 7 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001631_080 |
0.1800038497 |
- |
- |
PREDICTED: importin-5-like [Populus euphratica] |
| 4 |
Hb_000175_040 |
0.1927597007 |
- |
- |
hypothetical protein JCGZ_06119 [Jatropha curcas] |
| 5 |
Hb_001141_400 |
0.1937112194 |
- |
- |
hypothetical protein B456_007G2854002, partial [Gossypium raimondii] |
| 6 |
Hb_028365_010 |
0.202833951 |
- |
- |
PREDICTED: uncharacterized protein LOC105631661 [Jatropha curcas] |
| 7 |
Hb_003756_010 |
0.2058897301 |
- |
- |
PREDICTED: uncharacterized protein LOC105642108 [Jatropha curcas] |
| 8 |
Hb_000445_330 |
0.2087630188 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000373_290 |
0.211363707 |
- |
- |
RecName: Full=Citrate-binding protein; Flags: Precursor [Hevea brasiliensis] |
| 10 |
Hb_001109_120 |
0.2142260291 |
- |
- |
PREDICTED: acyl-CoA-binding protein-like [Nelumbo nucifera] |
| 11 |
Hb_005450_010 |
0.2166067146 |
- |
- |
PREDICTED: RNA-binding protein 24-B isoform X1 [Jatropha curcas] |
| 12 |
Hb_005410_030 |
0.2171958241 |
- |
- |
glutaredoxin 2 [Hevea brasiliensis] |
| 13 |
Hb_003057_130 |
0.2209691288 |
- |
- |
- |
| 14 |
Hb_000788_120 |
0.2236113567 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPS2-like [Eucalyptus grandis] |
| 15 |
Hb_004060_010 |
0.2276163248 |
- |
- |
- |
| 16 |
Hb_000086_330 |
0.2292751643 |
- |
- |
PREDICTED: glutathione gamma-glutamylcysteinyltransferase 1 [Jatropha curcas] |
| 17 |
Hb_001741_050 |
0.2335909085 |
- |
- |
- |
| 18 |
Hb_012395_100 |
0.234212423 |
- |
- |
eukaryotic translation initiation factor 4e, putative [Ricinus communis] |
| 19 |
Hb_003745_010 |
0.2351344142 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_03155 [Jatropha curcas] |
| 20 |
Hb_019516_040 |
0.2358700239 |
desease resistance |
Gene Name: AAA |
Katanin p60 ATPase-containing subunit, putative [Ricinus communis] |