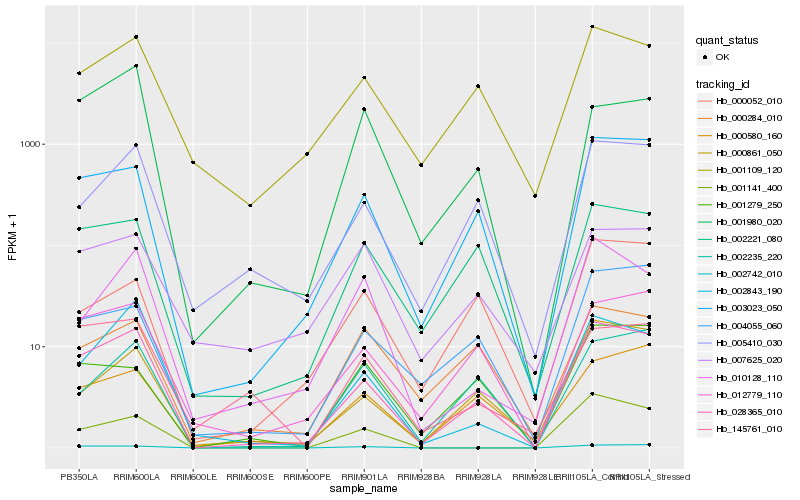
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_028365_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631661 [Jatropha curcas] |
| 2 |
Hb_003023_050 |
0.1247956974 |
- |
- |
hypothetical protein POPTR_0013s10320g [Populus trichocarpa] |
| 3 |
Hb_012779_110 |
0.145850081 |
- |
- |
PREDICTED: uncharacterized protein LOC105650717 [Jatropha curcas] |
| 4 |
Hb_005410_030 |
0.1548871209 |
- |
- |
glutaredoxin 2 [Hevea brasiliensis] |
| 5 |
Hb_002843_190 |
0.1605496704 |
transcription factor |
TF Family: GRF |
PREDICTED: growth-regulating factor 7 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001109_120 |
0.1644304191 |
- |
- |
PREDICTED: acyl-CoA-binding protein-like [Nelumbo nucifera] |
| 7 |
Hb_002221_080 |
0.1647781857 |
- |
- |
PREDICTED: uncharacterized protein LOC105639992 [Jatropha curcas] |
| 8 |
Hb_000861_050 |
0.1647841333 |
- |
- |
PREDICTED: transcription elongation factor SPT6-like isoform X1 [Populus euphratica] |
| 9 |
Hb_000580_160 |
0.1782583862 |
- |
- |
Nitrate transporter 1.5 [Morus notabilis] |
| 10 |
Hb_001141_400 |
0.1864791828 |
- |
- |
hypothetical protein B456_007G2854002, partial [Gossypium raimondii] |
| 11 |
Hb_004055_060 |
0.1870767922 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 5 [Jatropha curcas] |
| 12 |
Hb_002235_220 |
0.1895336907 |
- |
- |
PREDICTED: pectinesterase 31-like [Jatropha curcas] |
| 13 |
Hb_002742_010 |
0.195551657 |
- |
- |
PREDICTED: uncharacterized protein LOC104115978 [Nicotiana tomentosiformis] |
| 14 |
Hb_145761_010 |
0.1976330785 |
- |
- |
- |
| 15 |
Hb_001980_020 |
0.2027170891 |
- |
- |
- |
| 16 |
Hb_010128_110 |
0.202833951 |
- |
- |
PREDICTED: uncharacterized protein LOC105632071 [Jatropha curcas] |
| 17 |
Hb_001279_250 |
0.2047531218 |
- |
- |
PREDICTED: pectate lyase-like [Jatropha curcas] |
| 18 |
Hb_000284_010 |
0.2047574935 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_04974 [Jatropha curcas] |
| 19 |
Hb_007625_020 |
0.2097756716 |
- |
- |
PREDICTED: casein kinase II subunit alpha-like [Sesamum indicum] |
| 20 |
Hb_000052_010 |
0.2105492346 |
- |
- |
hypothetical protein M569_08416, partial [Genlisea aurea] |