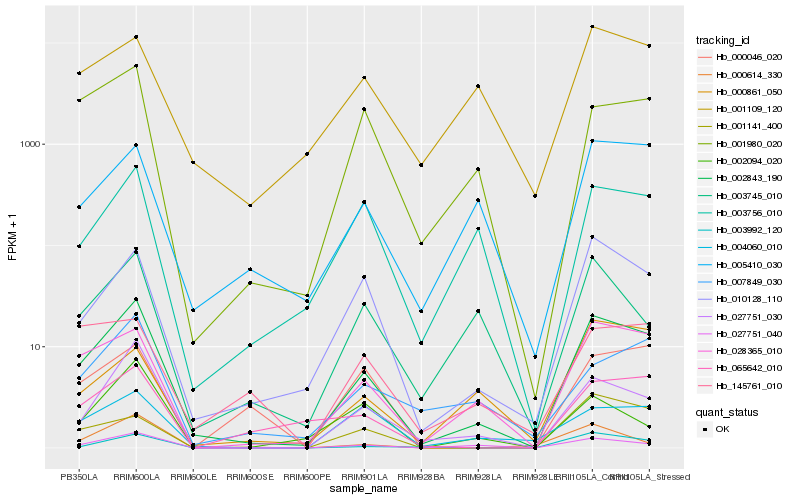
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002843_190 |
0.0 |
transcription factor |
TF Family: GRF |
PREDICTED: growth-regulating factor 7 isoform X1 [Jatropha curcas] |
| 2 |
Hb_028365_010 |
0.1605496704 |
- |
- |
PREDICTED: uncharacterized protein LOC105631661 [Jatropha curcas] |
| 3 |
Hb_027751_040 |
0.1638034364 |
- |
- |
PREDICTED: uncharacterized protein LOC105637052 [Jatropha curcas] |
| 4 |
Hb_027751_030 |
0.1735290058 |
- |
- |
- |
| 5 |
Hb_010128_110 |
0.1767079858 |
- |
- |
PREDICTED: uncharacterized protein LOC105632071 [Jatropha curcas] |
| 6 |
Hb_003992_120 |
0.2030502916 |
- |
- |
hypothetical protein MTR_6g089560 [Medicago truncatula] |
| 7 |
Hb_001980_020 |
0.2071212041 |
- |
- |
- |
| 8 |
Hb_005410_030 |
0.211313491 |
- |
- |
glutaredoxin 2 [Hevea brasiliensis] |
| 9 |
Hb_003756_010 |
0.2183845056 |
- |
- |
PREDICTED: uncharacterized protein LOC105642108 [Jatropha curcas] |
| 10 |
Hb_065642_010 |
0.2210112362 |
- |
- |
- |
| 11 |
Hb_007849_030 |
0.2236661257 |
- |
- |
l-allo-threonine aldolase, putative [Ricinus communis] |
| 12 |
Hb_004060_010 |
0.2247833398 |
- |
- |
- |
| 13 |
Hb_001109_120 |
0.2316477124 |
- |
- |
PREDICTED: acyl-CoA-binding protein-like [Nelumbo nucifera] |
| 14 |
Hb_001141_400 |
0.2347876376 |
- |
- |
hypothetical protein B456_007G2854002, partial [Gossypium raimondii] |
| 15 |
Hb_003745_010 |
0.2368186788 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_03155 [Jatropha curcas] |
| 16 |
Hb_000046_020 |
0.2396407446 |
- |
- |
- |
| 17 |
Hb_000614_330 |
0.2397837543 |
- |
- |
hypothetical protein POPTR_0008s18070g [Populus trichocarpa] |
| 18 |
Hb_002094_020 |
0.2457641935 |
- |
- |
PREDICTED: alpha-copaene synthase-like [Jatropha curcas] |
| 19 |
Hb_145761_010 |
0.2492650311 |
- |
- |
- |
| 20 |
Hb_000861_050 |
0.2498828763 |
- |
- |
PREDICTED: transcription elongation factor SPT6-like isoform X1 [Populus euphratica] |