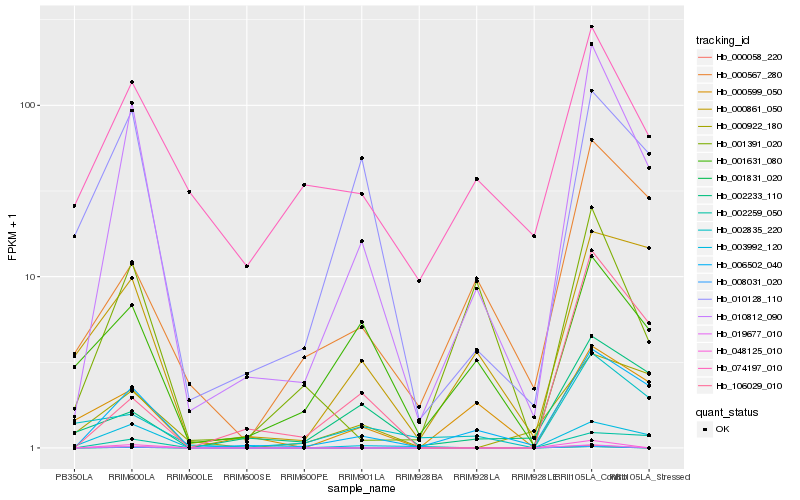
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010812_090 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase AtPK2/AtPK19-like [Jatropha curcas] |
| 2 |
Hb_006502_040 |
0.1623336609 |
transcription factor |
TF Family: bHLH |
Transcription factor ICE1, putative [Ricinus communis] |
| 3 |
Hb_003992_120 |
0.2312570499 |
- |
- |
hypothetical protein MTR_6g089560 [Medicago truncatula] |
| 4 |
Hb_010128_110 |
0.2583805613 |
- |
- |
PREDICTED: uncharacterized protein LOC105632071 [Jatropha curcas] |
| 5 |
Hb_000567_280 |
0.2831366431 |
- |
- |
PREDICTED: uncharacterized protein At4g06598-like [Populus euphratica] |
| 6 |
Hb_000861_050 |
0.2851274243 |
- |
- |
PREDICTED: transcription elongation factor SPT6-like isoform X1 [Populus euphratica] |
| 7 |
Hb_074197_010 |
0.286989896 |
- |
- |
PREDICTED: methylsterol monooxygenase 1-1-like [Jatropha curcas] |
| 8 |
Hb_001391_020 |
0.2881060928 |
- |
- |
Arylacetamide deacetylase, putative [Ricinus communis] |
| 9 |
Hb_002835_220 |
0.2894844909 |
- |
- |
PREDICTED: MATE efflux family protein 7 [Vitis vinifera] |
| 10 |
Hb_001631_080 |
0.2924470766 |
- |
- |
PREDICTED: importin-5-like [Populus euphratica] |
| 11 |
Hb_002259_050 |
0.2961473627 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 12 |
Hb_106029_010 |
0.2984387449 |
- |
- |
Transcription factor ICE1, putative [Ricinus communis] |
| 13 |
Hb_000922_180 |
0.2991404784 |
- |
- |
PREDICTED: probable inactive poly [ADP-ribose] polymerase SRO5 [Jatropha curcas] |
| 14 |
Hb_000599_050 |
0.3012041185 |
- |
- |
vacuolar ATP synthase subunit G plant, putative [Ricinus communis] |
| 15 |
Hb_002233_110 |
0.3029777114 |
- |
- |
PREDICTED: nuclear transcription factor Y subunit B-3-like [Jatropha curcas] |
| 16 |
Hb_008031_020 |
0.3059077427 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 17 |
Hb_001831_020 |
0.3059154695 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 18 |
Hb_000058_220 |
0.3059321493 |
- |
- |
hypothetical protein, partial [Staphylococcus aureus] |
| 19 |
Hb_019677_010 |
0.3059383776 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_048125_010 |
0.3060226021 |
- |
- |
PREDICTED: uncharacterized protein LOC104104928 [Nicotiana tomentosiformis] |