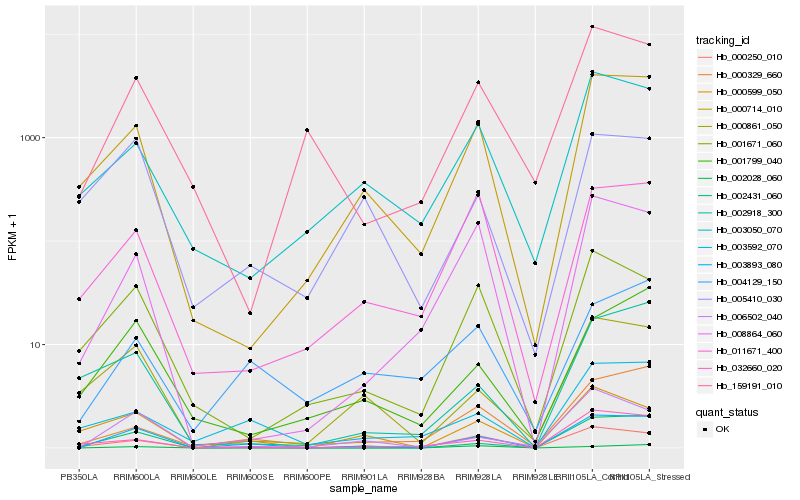
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003893_080 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_006502_040 |
0.1922101425 |
transcription factor |
TF Family: bHLH |
Transcription factor ICE1, putative [Ricinus communis] |
| 3 |
Hb_002431_060 |
0.2001016159 |
- |
- |
- |
| 4 |
Hb_000714_010 |
0.2061942596 |
- |
- |
PREDICTED: salt stress-induced hydrophobic peptide ESI3 [Vitis vinifera] |
| 5 |
Hb_000861_050 |
0.225897211 |
- |
- |
PREDICTED: transcription elongation factor SPT6-like isoform X1 [Populus euphratica] |
| 6 |
Hb_000599_050 |
0.2289883057 |
- |
- |
vacuolar ATP synthase subunit G plant, putative [Ricinus communis] |
| 7 |
Hb_003592_070 |
0.2292084167 |
- |
- |
- |
| 8 |
Hb_000250_010 |
0.2303843154 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 9 |
Hb_032660_020 |
0.23342382 |
- |
- |
PREDICTED: F-box protein At5g49610-like [Jatropha curcas] |
| 10 |
Hb_000329_660 |
0.2354309045 |
- |
- |
PREDICTED: carotenoid cleavage dioxygenase 7, chloroplastic [Jatropha curcas] |
| 11 |
Hb_008864_060 |
0.2369916827 |
- |
- |
PREDICTED: uncharacterized protein LOC105634361 [Jatropha curcas] |
| 12 |
Hb_001799_040 |
0.2432297321 |
- |
- |
- |
| 13 |
Hb_002028_060 |
0.2484817696 |
- |
- |
- |
| 14 |
Hb_003050_070 |
0.2491685067 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_159191_010 |
0.2504539177 |
- |
- |
protease inhibitor protein [Hevea brasiliensis] |
| 16 |
Hb_001671_060 |
0.2512907839 |
- |
- |
fad NAD binding oxidoreductases, putative [Ricinus communis] |
| 17 |
Hb_002918_300 |
0.2530401736 |
- |
- |
- |
| 18 |
Hb_004129_150 |
0.2533349236 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-4a-like [Jatropha curcas] |
| 19 |
Hb_005410_030 |
0.2622987653 |
- |
- |
glutaredoxin 2 [Hevea brasiliensis] |
| 20 |
Hb_011671_400 |
0.2650646794 |
- |
- |
PREDICTED: cyclin-U4-1 [Jatropha curcas] |