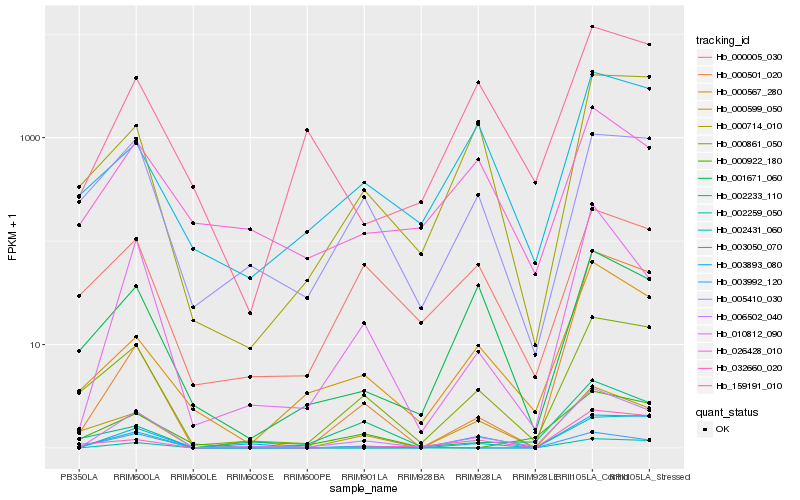
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006502_040 |
0.0 |
transcription factor |
TF Family: bHLH |
Transcription factor ICE1, putative [Ricinus communis] |
| 2 |
Hb_010812_090 |
0.1623336609 |
- |
- |
PREDICTED: serine/threonine-protein kinase AtPK2/AtPK19-like [Jatropha curcas] |
| 3 |
Hb_000861_050 |
0.1912719108 |
- |
- |
PREDICTED: transcription elongation factor SPT6-like isoform X1 [Populus euphratica] |
| 4 |
Hb_003893_080 |
0.1922101425 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000567_280 |
0.2071917911 |
- |
- |
PREDICTED: uncharacterized protein At4g06598-like [Populus euphratica] |
| 6 |
Hb_159191_010 |
0.2340558772 |
- |
- |
protease inhibitor protein [Hevea brasiliensis] |
| 7 |
Hb_002259_050 |
0.2368996521 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 8 |
Hb_002431_060 |
0.2370768595 |
- |
- |
- |
| 9 |
Hb_000714_010 |
0.2406997511 |
- |
- |
PREDICTED: salt stress-induced hydrophobic peptide ESI3 [Vitis vinifera] |
| 10 |
Hb_000599_050 |
0.2408706354 |
- |
- |
vacuolar ATP synthase subunit G plant, putative [Ricinus communis] |
| 11 |
Hb_003050_070 |
0.2455227317 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001671_060 |
0.2455897952 |
- |
- |
fad NAD binding oxidoreductases, putative [Ricinus communis] |
| 13 |
Hb_005410_030 |
0.2499454411 |
- |
- |
glutaredoxin 2 [Hevea brasiliensis] |
| 14 |
Hb_026428_010 |
0.2504001975 |
- |
- |
copper transport protein ATOX1 [Hevea brasiliensis] |
| 15 |
Hb_000005_030 |
0.2514773032 |
- |
- |
hypothetical protein POPTR_0009s00870g [Populus trichocarpa] |
| 16 |
Hb_003992_120 |
0.2529132933 |
- |
- |
hypothetical protein MTR_6g089560 [Medicago truncatula] |
| 17 |
Hb_002233_110 |
0.2567554807 |
- |
- |
PREDICTED: nuclear transcription factor Y subunit B-3-like [Jatropha curcas] |
| 18 |
Hb_000922_180 |
0.2588966311 |
- |
- |
PREDICTED: probable inactive poly [ADP-ribose] polymerase SRO5 [Jatropha curcas] |
| 19 |
Hb_000501_020 |
0.2621641458 |
- |
- |
PREDICTED: uncharacterized protein LOC100250168 [Vitis vinifera] |
| 20 |
Hb_032660_020 |
0.2628034066 |
- |
- |
PREDICTED: F-box protein At5g49610-like [Jatropha curcas] |