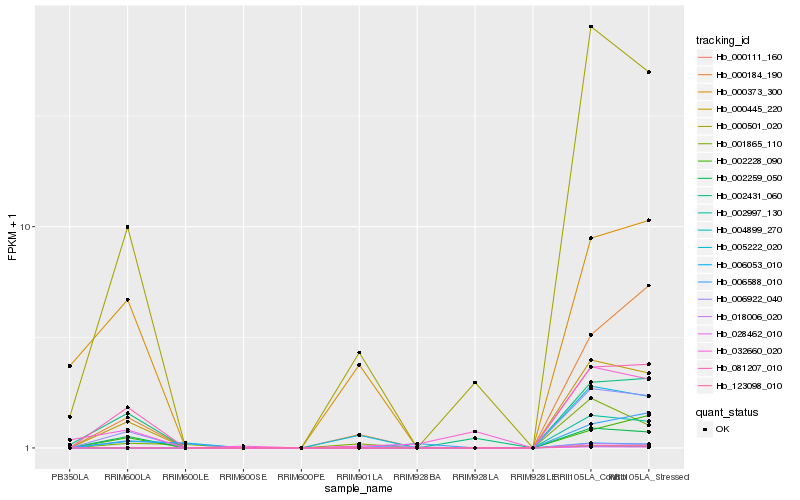
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006922_040 |
0.0 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 2 |
Hb_000445_220 |
0.0801119461 |
- |
- |
- |
| 3 |
Hb_081207_010 |
0.1139334025 |
- |
- |
hypothetical protein JCGZ_17195 [Jatropha curcas] |
| 4 |
Hb_000501_020 |
0.1258783733 |
- |
- |
PREDICTED: uncharacterized protein LOC100250168 [Vitis vinifera] |
| 5 |
Hb_002259_050 |
0.1682308742 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_002228_090 |
0.1917535818 |
- |
- |
hypothetical protein POPTR_0006s23560g [Populus trichocarpa] |
| 7 |
Hb_000184_190 |
0.2012809957 |
- |
- |
hypothetical protein VITISV_016970 [Vitis vinifera] |
| 8 |
Hb_002431_060 |
0.2060197758 |
- |
- |
- |
| 9 |
Hb_005222_020 |
0.230601641 |
- |
- |
hypothetical protein [Beta vulgaris subsp. vulgaris] |
| 10 |
Hb_032660_020 |
0.2351562683 |
- |
- |
PREDICTED: F-box protein At5g49610-like [Jatropha curcas] |
| 11 |
Hb_001865_110 |
0.2435587892 |
- |
- |
- |
| 12 |
Hb_000373_300 |
0.2464567436 |
- |
- |
hypothetical protein F775_20382 [Aegilops tauschii] |
| 13 |
Hb_006588_010 |
0.2468366799 |
- |
- |
- |
| 14 |
Hb_002997_130 |
0.2479476736 |
- |
- |
- |
| 15 |
Hb_028462_010 |
0.2479681303 |
- |
- |
PREDICTED: uncharacterized protein LOC101509514 [Cicer arietinum] |
| 16 |
Hb_123098_010 |
0.2479682842 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 17 |
Hb_004899_270 |
0.247969454 |
- |
- |
PREDICTED: beta-galactosidase 15-like [Cucumis sativus] |
| 18 |
Hb_000111_160 |
0.2479710422 |
- |
- |
Disease resistance family protein / LRR family protein, putative [Theobroma cacao] |
| 19 |
Hb_018006_020 |
0.2479745524 |
- |
- |
PREDICTED: uncharacterized protein LOC105641417 [Jatropha curcas] |
| 20 |
Hb_006053_010 |
0.2479755396 |
- |
- |
PREDICTED: uncharacterized protein LOC104216022, partial [Nicotiana sylvestris] |