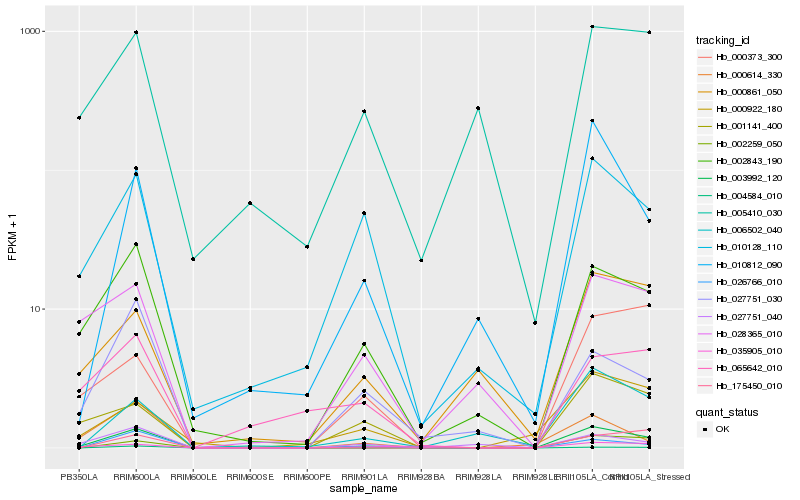
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003992_120 |
0.0 |
- |
- |
hypothetical protein MTR_6g089560 [Medicago truncatula] |
| 2 |
Hb_002843_190 |
0.2030502916 |
transcription factor |
TF Family: GRF |
PREDICTED: growth-regulating factor 7 isoform X1 [Jatropha curcas] |
| 3 |
Hb_010812_090 |
0.2312570499 |
- |
- |
PREDICTED: serine/threonine-protein kinase AtPK2/AtPK19-like [Jatropha curcas] |
| 4 |
Hb_035905_010 |
0.2358614635 |
- |
- |
PREDICTED: uncharacterized protein LOC105627956 [Jatropha curcas] |
| 5 |
Hb_010128_110 |
0.2396088641 |
- |
- |
PREDICTED: uncharacterized protein LOC105632071 [Jatropha curcas] |
| 6 |
Hb_001141_400 |
0.243564503 |
- |
- |
hypothetical protein B456_007G2854002, partial [Gossypium raimondii] |
| 7 |
Hb_026766_010 |
0.2440557866 |
- |
- |
- |
| 8 |
Hb_002259_050 |
0.2478992431 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 9 |
Hb_006502_040 |
0.2529132933 |
transcription factor |
TF Family: bHLH |
Transcription factor ICE1, putative [Ricinus communis] |
| 10 |
Hb_027751_030 |
0.2540888344 |
- |
- |
- |
| 11 |
Hb_000922_180 |
0.2614476275 |
- |
- |
PREDICTED: probable inactive poly [ADP-ribose] polymerase SRO5 [Jatropha curcas] |
| 12 |
Hb_028365_010 |
0.2665854327 |
- |
- |
PREDICTED: uncharacterized protein LOC105631661 [Jatropha curcas] |
| 13 |
Hb_000861_050 |
0.2686770866 |
- |
- |
PREDICTED: transcription elongation factor SPT6-like isoform X1 [Populus euphratica] |
| 14 |
Hb_004584_010 |
0.2717483387 |
- |
- |
PREDICTED: uncharacterized protein LOC104094989 [Nicotiana tomentosiformis] |
| 15 |
Hb_027751_040 |
0.2785813073 |
- |
- |
PREDICTED: uncharacterized protein LOC105637052 [Jatropha curcas] |
| 16 |
Hb_000614_330 |
0.2788274098 |
- |
- |
hypothetical protein POPTR_0008s18070g [Populus trichocarpa] |
| 17 |
Hb_000373_300 |
0.2852852781 |
- |
- |
hypothetical protein F775_20382 [Aegilops tauschii] |
| 18 |
Hb_005410_030 |
0.2880607051 |
- |
- |
glutaredoxin 2 [Hevea brasiliensis] |
| 19 |
Hb_175450_010 |
0.2881984206 |
- |
- |
- |
| 20 |
Hb_065642_010 |
0.2983077196 |
- |
- |
- |