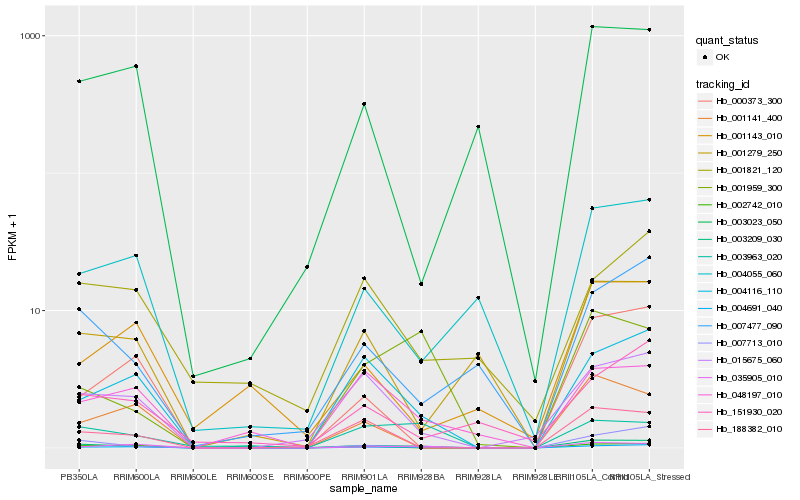
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004691_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105775124 [Gossypium raimondii] |
| 2 |
Hb_004116_110 |
0.1463586954 |
- |
- |
hypothetical protein MTR_3g064100 [Medicago truncatula] |
| 3 |
Hb_035905_010 |
0.1599507672 |
- |
- |
PREDICTED: uncharacterized protein LOC105627956 [Jatropha curcas] |
| 4 |
Hb_015675_060 |
0.2062973591 |
- |
- |
- |
| 5 |
Hb_002742_010 |
0.2217277558 |
- |
- |
PREDICTED: uncharacterized protein LOC104115978 [Nicotiana tomentosiformis] |
| 6 |
Hb_000373_300 |
0.2250420686 |
- |
- |
hypothetical protein F775_20382 [Aegilops tauschii] |
| 7 |
Hb_004055_060 |
0.2260564035 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 5 [Jatropha curcas] |
| 8 |
Hb_048197_010 |
0.2317086449 |
- |
- |
- |
| 9 |
Hb_007713_010 |
0.2348820996 |
- |
- |
hypothetical protein JCGZ_18011 [Jatropha curcas] |
| 10 |
Hb_151930_020 |
0.2418949121 |
- |
- |
Wound-induced protein WIN1 precursor, putative [Ricinus communis] |
| 11 |
Hb_003209_030 |
0.2451553707 |
- |
- |
PREDICTED: heparanase-like protein 2 [Jatropha curcas] |
| 12 |
Hb_007477_090 |
0.2499512037 |
- |
- |
PREDICTED: tetraspanin-18 [Jatropha curcas] |
| 13 |
Hb_001821_120 |
0.2528224265 |
- |
- |
PREDICTED: uncharacterized protein LOC105643956 [Jatropha curcas] |
| 14 |
Hb_003963_020 |
0.2544538331 |
- |
- |
PREDICTED: uncharacterized protein LOC100256672 [Vitis vinifera] |
| 15 |
Hb_001141_400 |
0.2581838304 |
- |
- |
hypothetical protein B456_007G2854002, partial [Gossypium raimondii] |
| 16 |
Hb_003023_050 |
0.2612967961 |
- |
- |
hypothetical protein POPTR_0013s10320g [Populus trichocarpa] |
| 17 |
Hb_001143_010 |
0.2672483949 |
- |
- |
- |
| 18 |
Hb_001279_250 |
0.267838546 |
- |
- |
PREDICTED: pectate lyase-like [Jatropha curcas] |
| 19 |
Hb_001959_300 |
0.269287576 |
- |
- |
PREDICTED: 3-isopropylmalate dehydratase large subunit [Jatropha curcas] |
| 20 |
Hb_188382_010 |
0.269758857 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |