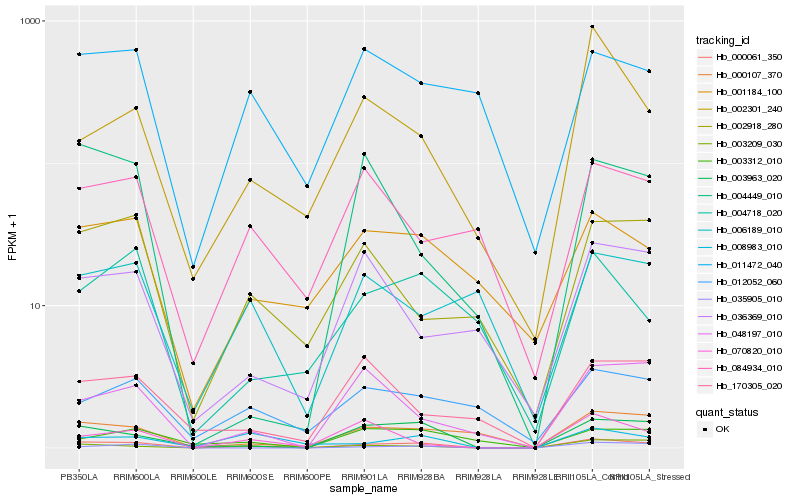
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000061_350 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_003963_020 |
0.1830213032 |
- |
- |
PREDICTED: uncharacterized protein LOC100256672 [Vitis vinifera] |
| 3 |
Hb_003209_030 |
0.2322852517 |
- |
- |
PREDICTED: heparanase-like protein 2 [Jatropha curcas] |
| 4 |
Hb_000107_370 |
0.2387654607 |
- |
- |
PREDICTED: uncharacterized protein At2g17340-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_048197_010 |
0.247819994 |
- |
- |
- |
| 6 |
Hb_035905_010 |
0.2511103034 |
- |
- |
PREDICTED: uncharacterized protein LOC105627956 [Jatropha curcas] |
| 7 |
Hb_002301_240 |
0.2526814275 |
- |
- |
auxin-repressed protein-like protein ARP1 [Manihot esculenta] |
| 8 |
Hb_008983_010 |
0.2570023608 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 9 |
Hb_002918_280 |
0.2576118465 |
- |
- |
hypothetical protein POPTR_0004s04800g [Populus trichocarpa] |
| 10 |
Hb_001184_100 |
0.2578847805 |
- |
- |
PREDICTED: zinc finger protein 4-like [Populus euphratica] |
| 11 |
Hb_070820_010 |
0.2601864162 |
- |
- |
hypothetical protein B456_008G232800 [Gossypium raimondii] |
| 12 |
Hb_004449_010 |
0.2650938846 |
- |
- |
lipase [Ricinus communis] |
| 13 |
Hb_004718_020 |
0.266623487 |
- |
- |
Potassium channel KAT3, putative [Ricinus communis] |
| 14 |
Hb_003312_010 |
0.2677520415 |
- |
- |
PREDICTED: uncharacterized protein LOC104428797 [Eucalyptus grandis] |
| 15 |
Hb_012052_060 |
0.2731967447 |
- |
- |
hypothetical protein POPTR_0018s10190g [Populus trichocarpa] |
| 16 |
Hb_084934_010 |
0.2735818477 |
- |
- |
hypothetical protein JCGZ_22225 [Jatropha curcas] |
| 17 |
Hb_011472_040 |
0.2736036047 |
- |
- |
PREDICTED: clathrin light chain 3 [Jatropha curcas] |
| 18 |
Hb_036369_010 |
0.2737270142 |
- |
- |
- |
| 19 |
Hb_006189_010 |
0.2785829106 |
- |
- |
- |
| 20 |
Hb_170305_020 |
0.2791200922 |
- |
- |
hypothetical protein JCGZ_23578 [Jatropha curcas] |