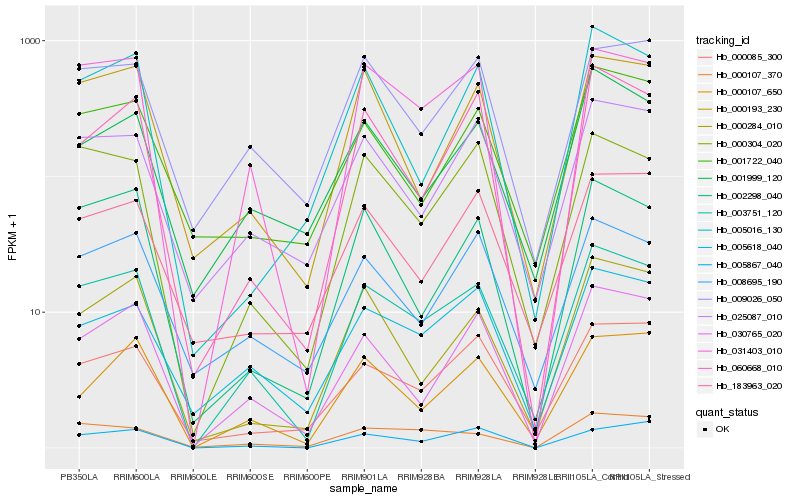
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003751_120 |
0.0 |
- |
- |
- |
| 2 |
Hb_031403_010 |
0.0901100171 |
- |
- |
hypothetical protein POPTR_0001s40140g [Populus trichocarpa] |
| 3 |
Hb_000304_020 |
0.1148074398 |
- |
- |
beta-1,3-glucanase form RRII Gln 3 [Hevea brasiliensis] |
| 4 |
Hb_030765_020 |
0.12185313 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 5 |
Hb_005618_040 |
0.1222326485 |
- |
- |
- |
| 6 |
Hb_025087_010 |
0.1238258289 |
- |
- |
PREDICTED: RING-H2 finger protein ATL56 [Jatropha curcas] |
| 7 |
Hb_000085_300 |
0.1272726728 |
- |
- |
- |
| 8 |
Hb_002298_040 |
0.1285103843 |
- |
- |
PREDICTED: uncharacterized protein LOC105633642 [Jatropha curcas] |
| 9 |
Hb_008695_190 |
0.1306461762 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 17 [Jatropha curcas] |
| 10 |
Hb_000284_010 |
0.130996639 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_04974 [Jatropha curcas] |
| 11 |
Hb_000193_230 |
0.1312170931 |
- |
- |
PREDICTED: NEP1-interacting protein-like 2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001999_120 |
0.1316165197 |
- |
- |
PREDICTED: pleckstrin homology domain-containing protein 1 [Jatropha curcas] |
| 13 |
Hb_060668_010 |
0.1337708831 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein indeterminate-domain 2-like [Jatropha curcas] |
| 14 |
Hb_009026_050 |
0.1338988823 |
- |
- |
vacuolar ATPase F subunit [Hevea brasiliensis] |
| 15 |
Hb_000107_370 |
0.1347277337 |
- |
- |
PREDICTED: uncharacterized protein At2g17340-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_000107_650 |
0.13760664 |
- |
- |
PREDICTED: cyclic phosphodiesterase-like [Jatropha curcas] |
| 17 |
Hb_183963_020 |
0.1380944588 |
- |
- |
PREDICTED: uncharacterized protein LOC105644615 [Jatropha curcas] |
| 18 |
Hb_001722_040 |
0.1388113059 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 variant 1A-like [Jatropha curcas] |
| 19 |
Hb_005867_040 |
0.1402733215 |
transcription factor |
TF Family: RWP-RK |
PREDICTED: protein RKD4 [Jatropha curcas] |
| 20 |
Hb_005016_130 |
0.1410528475 |
- |
- |
PREDICTED: uncharacterized protein LOC105642243 [Jatropha curcas] |