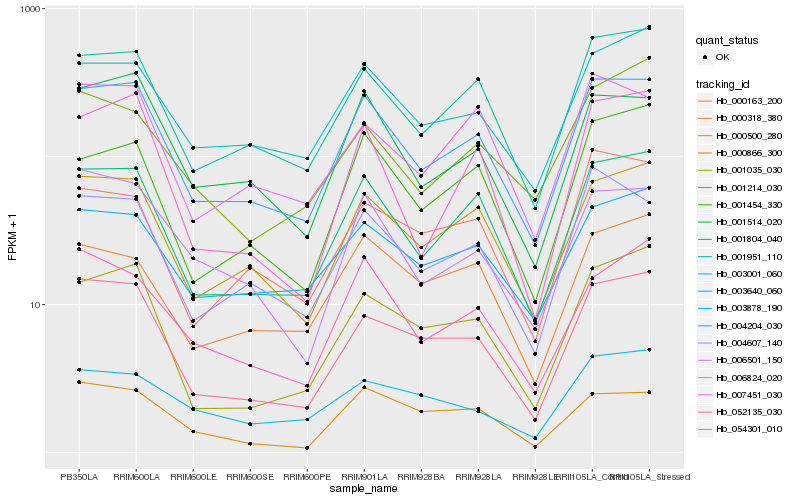
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_054301_010 |
0.0 |
- |
- |
hypothetical protein EUTSA_v10006737mg [Eutrema salsugineum] |
| 2 |
Hb_001035_030 |
0.0790914451 |
- |
- |
PREDICTED: probable rhamnogalacturonate lyase B isoform X1 [Jatropha curcas] |
| 3 |
Hb_004204_030 |
0.0893226025 |
- |
- |
unknown [Populus trichocarpa] |
| 4 |
Hb_000866_300 |
0.0943425133 |
- |
- |
hypothetical protein CICLE_v10023145mg [Citrus clementina] |
| 5 |
Hb_001804_040 |
0.1010972199 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001951_110 |
0.1048918159 |
- |
- |
PREDICTED: DNA-binding protein DDB_G0278111 [Jatropha curcas] |
| 7 |
Hb_003001_060 |
0.1250129351 |
- |
- |
wound-responsive family protein [Populus trichocarpa] |
| 8 |
Hb_000163_200 |
0.1250948005 |
- |
- |
PREDICTED: uncharacterized protein LOC105642527 [Jatropha curcas] |
| 9 |
Hb_000500_280 |
0.1262440504 |
- |
- |
Brassinosteroid-regulated protein BRU1 precursor, putative [Ricinus communis] |
| 10 |
Hb_003878_190 |
0.1268489914 |
- |
- |
hypothetical protein POPTR_0011s11040g [Populus trichocarpa] |
| 11 |
Hb_003640_060 |
0.12869664 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 12 |
Hb_004607_140 |
0.1319040496 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor PLT1 [Populus euphratica] |
| 13 |
Hb_052135_030 |
0.1337658515 |
- |
- |
PREDICTED: huntingtin-interacting protein K [Jatropha curcas] |
| 14 |
Hb_006824_020 |
0.1357441548 |
- |
- |
PREDICTED: thioredoxin domain-containing protein 9 homolog [Jatropha curcas] |
| 15 |
Hb_001214_030 |
0.1364752235 |
- |
- |
hypothetical protein POPTR_0006s10280g [Populus trichocarpa] |
| 16 |
Hb_006501_150 |
0.1366915822 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_007451_030 |
0.1370261295 |
- |
- |
PREDICTED: mitochondrial thiamine pyrophosphate carrier isoform X1 [Jatropha curcas] |
| 18 |
Hb_000318_380 |
0.1381139281 |
- |
- |
PREDICTED: diphthamide biosynthesis protein 3-like [Jatropha curcas] |
| 19 |
Hb_001454_330 |
0.1395735993 |
- |
- |
PREDICTED: 60S ribosomal protein L22-2-like [Jatropha curcas] |
| 20 |
Hb_001514_020 |
0.1405760904 |
- |
- |
PREDICTED: protein DCL, chloroplastic-like [Jatropha curcas] |