| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
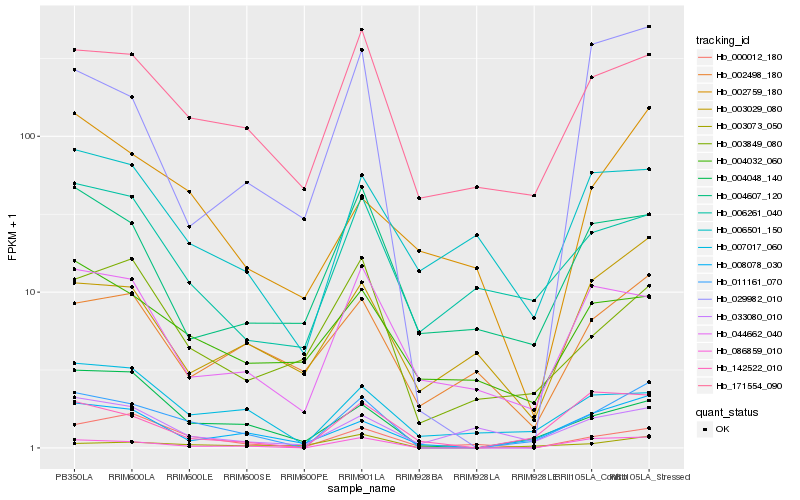
Hb_011161_070 |
0.0 |
- |
- |
PREDICTED: kynurenine--oxoglutarate transaminase 1-like isoform X1 [Populus euphratica] |
| 2 |
Hb_086859_010 |
0.169082827 |
- |
- |
- |
| 3 |
Hb_008078_030 |
0.1865018598 |
- |
- |
- |
| 4 |
Hb_142522_010 |
0.2103743335 |
- |
- |
- |
| 5 |
Hb_171554_090 |
0.211065703 |
- |
- |
PREDICTED: 10 kDa chaperonin [Jatropha curcas] |
| 6 |
Hb_004048_140 |
0.2213319908 |
- |
- |
- |
| 7 |
Hb_003073_050 |
0.231720677 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os03g0620400-like [Jatropha curcas] |
| 8 |
Hb_029982_010 |
0.233030486 |
- |
- |
hypothetical protein CICLE_v10006145mg [Citrus clementina] |
| 9 |
Hb_007017_060 |
0.2332606007 |
- |
- |
PREDICTED: 5-formyltetrahydrofolate cyclo-ligase, mitochondrial-like [Jatropha curcas] |
| 10 |
Hb_003849_080 |
0.2426809364 |
- |
- |
PREDICTED: sufE-like protein 2, chloroplastic isoform X2 [Jatropha curcas] |
| 11 |
Hb_000012_180 |
0.2427542327 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_044662_040 |
0.2430259254 |
- |
- |
- |
| 13 |
Hb_003029_080 |
0.2478391639 |
- |
- |
peptidyl-prolyl cis-trans isomerase, putative [Ricinus communis] |
| 14 |
Hb_006261_040 |
0.2479346392 |
- |
- |
PREDICTED: uncharacterized protein LOC105646602 [Jatropha curcas] |
| 15 |
Hb_002759_180 |
0.2483065091 |
- |
- |
PREDICTED: snakin-2-like [Jatropha curcas] |
| 16 |
Hb_004607_120 |
0.2513938228 |
- |
- |
PREDICTED: zinc finger A20 and AN1 domain-containing stress-associated protein 8 [Jatropha curcas] |
| 17 |
Hb_006501_150 |
0.2530822417 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002498_180 |
0.2534275928 |
- |
- |
PREDICTED: U3 small nucleolar RNA-associated protein 6 homolog [Jatropha curcas] |
| 19 |
Hb_033080_010 |
0.2536340543 |
- |
- |
- |
| 20 |
Hb_004032_060 |
0.2544601588 |
- |
- |
elongation factor 1 gamma-like protein, partial [Ipomoea nil] |