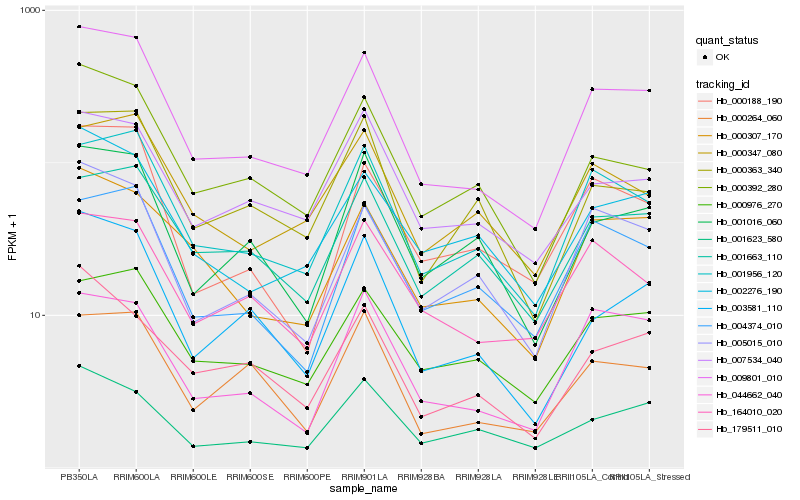
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009801_010 |
0.0 |
- |
- |
ATPase inhibitor, putative [Ricinus communis] |
| 2 |
Hb_001956_120 |
0.0906979797 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_005015_010 |
0.0995950741 |
- |
- |
PREDICTED: programmed cell death protein 2-like [Jatropha curcas] |
| 4 |
Hb_000392_280 |
0.1042333462 |
- |
- |
PREDICTED: pectinesterase 3 [Jatropha curcas] |
| 5 |
Hb_000307_170 |
0.1046422255 |
- |
- |
PREDICTED: uncharacterized protein LOC105638528 [Jatropha curcas] |
| 6 |
Hb_000976_270 |
0.1075246632 |
- |
- |
PREDICTED: uncharacterized protein LOC105646226 [Jatropha curcas] |
| 7 |
Hb_003581_110 |
0.1089162368 |
- |
- |
PREDICTED: uncharacterized protein LOC105649778 isoform X2 [Jatropha curcas] |
| 8 |
Hb_002276_190 |
0.1100212109 |
- |
- |
PREDICTED: uncharacterized protein LOC105640686 [Jatropha curcas] |
| 9 |
Hb_000188_190 |
0.1142632954 |
- |
- |
- |
| 10 |
Hb_001016_060 |
0.1165257483 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_164010_020 |
0.1171534236 |
- |
- |
PREDICTED: DNA-directed RNA polymerases I and III subunit RPAC1 [Jatropha curcas] |
| 12 |
Hb_007534_040 |
0.1179322443 |
- |
- |
PREDICTED: uncharacterized protein LOC105635538 [Jatropha curcas] |
| 13 |
Hb_004374_010 |
0.119452138 |
- |
- |
PREDICTED: PIN2/TERF1-interacting telomerase inhibitor 1 [Jatropha curcas] |
| 14 |
Hb_044662_040 |
0.1204099279 |
- |
- |
- |
| 15 |
Hb_000347_080 |
0.1208211266 |
- |
- |
PREDICTED: 25.3 kDa vesicle transport protein [Jatropha curcas] |
| 16 |
Hb_179511_010 |
0.1209226026 |
- |
- |
- |
| 17 |
Hb_001663_110 |
0.1210846657 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 18 |
Hb_000363_340 |
0.1213888687 |
- |
- |
PREDICTED: syntaxin-52-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_001623_580 |
0.1218462551 |
- |
- |
PREDICTED: histone-lysine N-methyltransferase family member SUVH9-like [Jatropha curcas] |
| 20 |
Hb_000264_060 |
0.121865565 |
- |
- |
PREDICTED: RNA polymerase-associated protein RTF1 homolog [Jatropha curcas] |