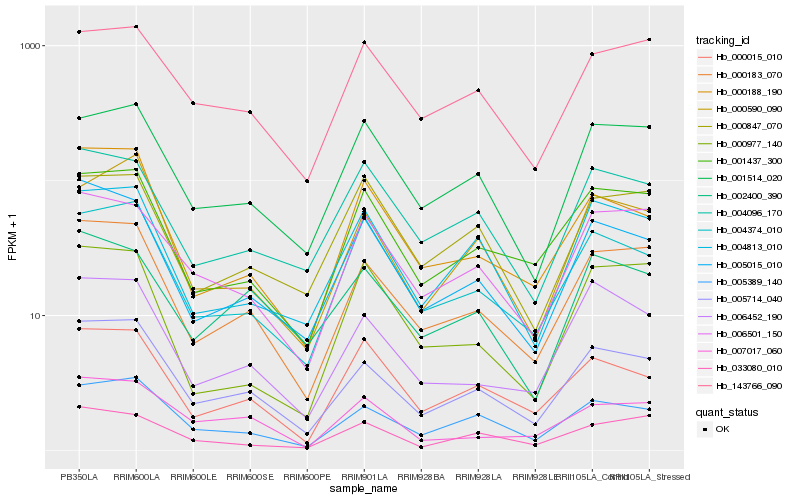
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000183_070 |
0.0 |
- |
- |
- |
| 2 |
Hb_005714_040 |
0.063721873 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001437_300 |
0.0926288135 |
- |
- |
- |
| 4 |
Hb_005015_010 |
0.0940961287 |
- |
- |
PREDICTED: programmed cell death protein 2-like [Jatropha curcas] |
| 5 |
Hb_006501_150 |
0.0958889035 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_006452_190 |
0.1013688415 |
- |
- |
PREDICTED: 3-isopropylmalate dehydrogenase 2, chloroplastic-like [Jatropha curcas] |
| 7 |
Hb_005389_140 |
0.1030793993 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase protein 14 precursor, putative [Ricinus communis] |
| 8 |
Hb_001514_020 |
0.1042599852 |
- |
- |
PREDICTED: protein DCL, chloroplastic-like [Jatropha curcas] |
| 9 |
Hb_143766_090 |
0.1054815521 |
- |
- |
60S ribosomal protein L31B [Hevea brasiliensis] |
| 10 |
Hb_000188_190 |
0.1134355253 |
- |
- |
- |
| 11 |
Hb_004374_010 |
0.1150644059 |
- |
- |
PREDICTED: PIN2/TERF1-interacting telomerase inhibitor 1 [Jatropha curcas] |
| 12 |
Hb_004813_010 |
0.1156044407 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 13 |
Hb_000977_140 |
0.1179174003 |
- |
- |
PREDICTED: uncharacterized protein LOC105801335 [Gossypium raimondii] |
| 14 |
Hb_033080_010 |
0.1199557355 |
- |
- |
- |
| 15 |
Hb_004096_170 |
0.121132606 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0275933 [Jatropha curcas] |
| 16 |
Hb_000590_090 |
0.121564019 |
- |
- |
PREDICTED: sulfate transporter 4.1, chloroplastic-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_000015_010 |
0.1230548228 |
- |
- |
- |
| 18 |
Hb_007017_060 |
0.123573895 |
- |
- |
PREDICTED: 5-formyltetrahydrofolate cyclo-ligase, mitochondrial-like [Jatropha curcas] |
| 19 |
Hb_000847_070 |
0.1248406103 |
- |
- |
PREDICTED: uncharacterized protein LOC105125699 [Populus euphratica] |
| 20 |
Hb_002400_390 |
0.1263905218 |
- |
- |
PREDICTED: 60S ribosomal protein L4 [Vitis vinifera] |