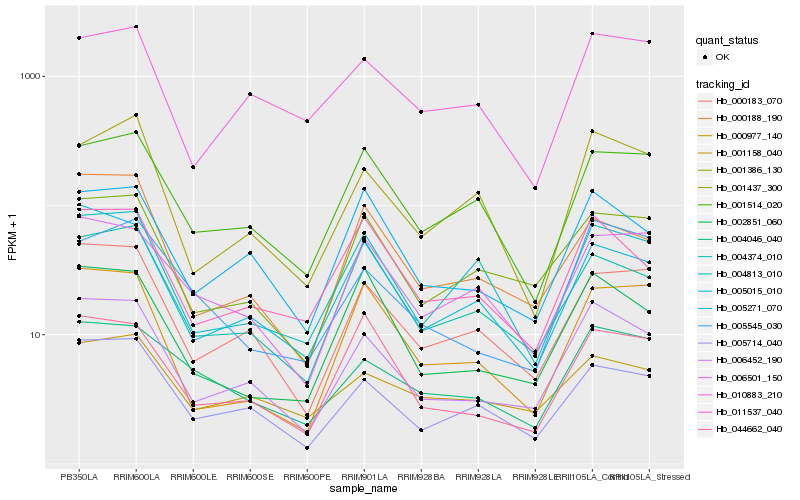
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006452_190 |
0.0 |
- |
- |
PREDICTED: 3-isopropylmalate dehydrogenase 2, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_000183_070 |
0.1013688415 |
- |
- |
- |
| 3 |
Hb_005714_040 |
0.1034642519 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001437_300 |
0.114322015 |
- |
- |
- |
| 5 |
Hb_005271_070 |
0.1150792341 |
- |
- |
PREDICTED: RNA-binding protein NOB1 [Jatropha curcas] |
| 6 |
Hb_005015_010 |
0.1178129914 |
- |
- |
PREDICTED: programmed cell death protein 2-like [Jatropha curcas] |
| 7 |
Hb_011537_040 |
0.1242853912 |
- |
- |
PREDICTED: uncharacterized protein LOC105646229 [Jatropha curcas] |
| 8 |
Hb_004374_010 |
0.1270058048 |
- |
- |
PREDICTED: PIN2/TERF1-interacting telomerase inhibitor 1 [Jatropha curcas] |
| 9 |
Hb_002851_060 |
0.1294037062 |
- |
- |
Clathrin light chain 2 -like protein [Gossypium arboreum] |
| 10 |
Hb_004046_040 |
0.1298901782 |
transcription factor |
TF Family: GRF |
PREDICTED: growth-regulating factor 4-like [Jatropha curcas] |
| 11 |
Hb_001386_130 |
0.1333178454 |
- |
- |
Mitochondrial import receptor subunit TOM20, putative [Ricinus communis] |
| 12 |
Hb_000188_190 |
0.1333801565 |
- |
- |
- |
| 13 |
Hb_044662_040 |
0.1340786898 |
- |
- |
- |
| 14 |
Hb_006501_150 |
0.1358074464 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_005545_030 |
0.1387457343 |
- |
- |
PREDICTED: uncharacterized protein LOC105645697 [Jatropha curcas] |
| 16 |
Hb_000977_140 |
0.1402433282 |
- |
- |
PREDICTED: uncharacterized protein LOC105801335 [Gossypium raimondii] |
| 17 |
Hb_001514_020 |
0.1406681594 |
- |
- |
PREDICTED: protein DCL, chloroplastic-like [Jatropha curcas] |
| 18 |
Hb_004813_010 |
0.1408914668 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 19 |
Hb_010883_210 |
0.1426545777 |
- |
- |
PREDICTED: selenoprotein K [Jatropha curcas] |
| 20 |
Hb_001158_040 |
0.1448394569 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3A isoform X2 [Jatropha curcas] |