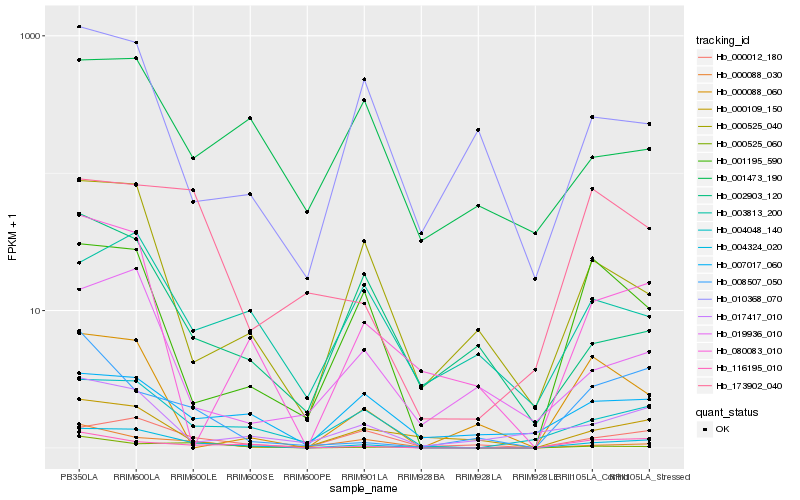
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004324_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_004048_140 |
0.1885101633 |
- |
- |
- |
| 3 |
Hb_000012_180 |
0.2394049276 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_116195_010 |
0.2399780767 |
- |
- |
- |
| 5 |
Hb_080083_010 |
0.2434544619 |
- |
- |
- |
| 6 |
Hb_000525_040 |
0.2499679292 |
- |
- |
- |
| 7 |
Hb_017417_010 |
0.2518794874 |
- |
- |
- |
| 8 |
Hb_000088_030 |
0.2568433583 |
- |
- |
hypothetical protein JCGZ_10947 [Jatropha curcas] |
| 9 |
Hb_019936_010 |
0.2579079233 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 10 [Jatropha curcas] |
| 10 |
Hb_001195_590 |
0.2598055401 |
- |
- |
PREDICTED: uncharacterized protein LOC105633753 [Jatropha curcas] |
| 11 |
Hb_000109_150 |
0.2630043971 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002903_120 |
0.2686316344 |
- |
- |
- |
| 13 |
Hb_007017_060 |
0.274203279 |
- |
- |
PREDICTED: 5-formyltetrahydrofolate cyclo-ligase, mitochondrial-like [Jatropha curcas] |
| 14 |
Hb_173902_040 |
0.2761268243 |
- |
- |
- |
| 15 |
Hb_008507_050 |
0.2805653445 |
- |
- |
PREDICTED: S-adenosylmethionine decarboxylase proenzyme-like [Jatropha curcas] |
| 16 |
Hb_003813_200 |
0.2810396868 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 17 |
Hb_010368_070 |
0.283409919 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000088_060 |
0.2838956762 |
- |
- |
- |
| 19 |
Hb_001473_190 |
0.2854392818 |
- |
- |
50S ribosomal protein L20 [Hevea brasiliensis] |
| 20 |
Hb_000525_060 |
0.2856929409 |
- |
- |
PREDICTED: uncharacterized protein LOC104211307 [Nicotiana sylvestris] |