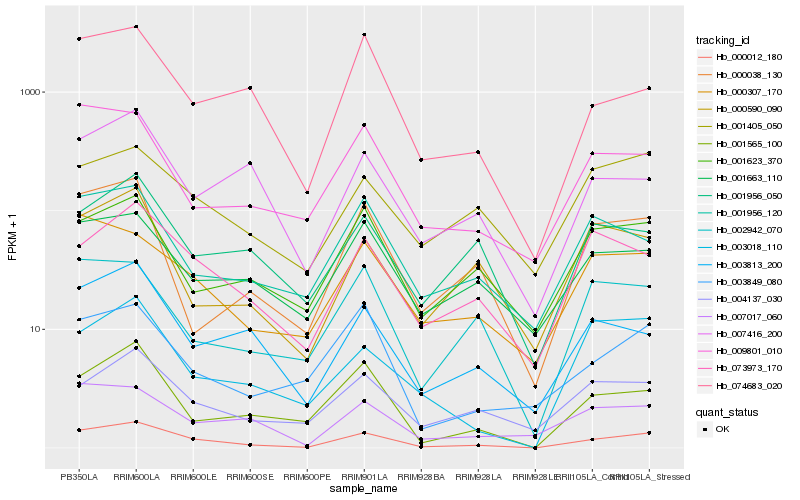
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000012_180 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001565_100 |
0.1648591821 |
- |
- |
unnamed protein product [Coffea canephora] |
| 3 |
Hb_073973_170 |
0.1716636061 |
- |
- |
PREDICTED: uncharacterized protein LOC105638918 [Jatropha curcas] |
| 4 |
Hb_074683_020 |
0.1732632374 |
- |
- |
PREDICTED: 60S ribosomal protein L27-3 [Jatropha curcas] |
| 5 |
Hb_003813_200 |
0.1758912191 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 6 |
Hb_000038_130 |
0.1804686152 |
- |
- |
- |
| 7 |
Hb_000307_170 |
0.1807974808 |
- |
- |
PREDICTED: uncharacterized protein LOC105638528 [Jatropha curcas] |
| 8 |
Hb_007017_060 |
0.1830207561 |
- |
- |
PREDICTED: 5-formyltetrahydrofolate cyclo-ligase, mitochondrial-like [Jatropha curcas] |
| 9 |
Hb_004137_030 |
0.1842798414 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_003849_080 |
0.1848304845 |
- |
- |
PREDICTED: sufE-like protein 2, chloroplastic isoform X2 [Jatropha curcas] |
| 11 |
Hb_002942_070 |
0.1850653569 |
- |
- |
PREDICTED: centromere protein S isoform X1 [Jatropha curcas] |
| 12 |
Hb_003018_110 |
0.1886901176 |
- |
- |
- |
| 13 |
Hb_001623_370 |
0.188732445 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_009801_010 |
0.1894096841 |
- |
- |
ATPase inhibitor, putative [Ricinus communis] |
| 15 |
Hb_007416_200 |
0.1918502964 |
- |
- |
PREDICTED: putative peptidyl-tRNA hydrolase PTRHD1 [Jatropha curcas] |
| 16 |
Hb_001663_110 |
0.1920210081 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 17 |
Hb_001405_050 |
0.1922692752 |
- |
- |
PREDICTED: uncharacterized protein LOC100500641 [Glycine max] |
| 18 |
Hb_001956_050 |
0.1923071298 |
- |
- |
PREDICTED: uncharacterized protein C20orf24 homolog [Jatropha curcas] |
| 19 |
Hb_001956_120 |
0.1942387907 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000590_090 |
0.194595421 |
- |
- |
PREDICTED: sulfate transporter 4.1, chloroplastic-like isoform X1 [Jatropha curcas] |