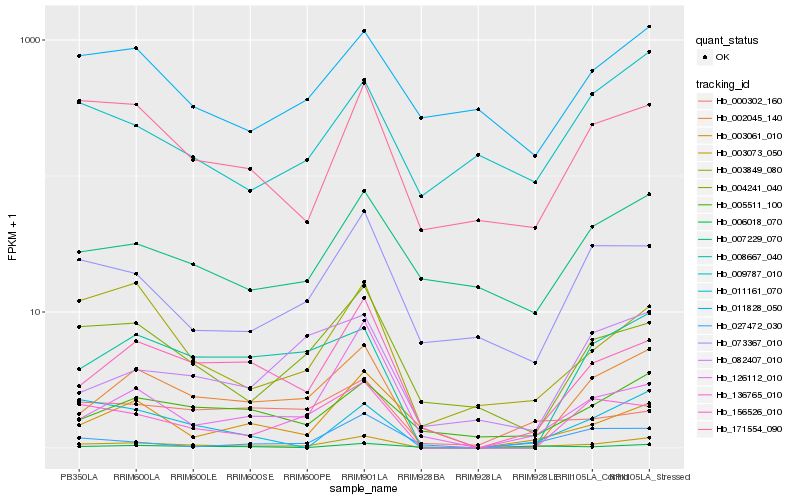
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003073_050 |
0.0 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os03g0620400-like [Jatropha curcas] |
| 2 |
Hb_002045_140 |
0.1808610433 |
- |
- |
histone acetyltransferase, partial [Triticum aestivum] |
| 3 |
Hb_156526_010 |
0.1871444718 |
- |
- |
PREDICTED: uncharacterized protein LOC105636715 [Jatropha curcas] |
| 4 |
Hb_006018_070 |
0.2087792142 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 5 |
Hb_005511_100 |
0.2090553696 |
- |
- |
PREDICTED: uncharacterized protein LOC105645493 isoform X1 [Jatropha curcas] |
| 6 |
Hb_171554_090 |
0.2191402678 |
- |
- |
PREDICTED: 10 kDa chaperonin [Jatropha curcas] |
| 7 |
Hb_004241_040 |
0.221174758 |
- |
- |
- |
| 8 |
Hb_003849_080 |
0.2211862211 |
- |
- |
PREDICTED: sufE-like protein 2, chloroplastic isoform X2 [Jatropha curcas] |
| 9 |
Hb_073367_010 |
0.2275482436 |
- |
- |
PREDICTED: origin of replication complex subunit 2 [Jatropha curcas] |
| 10 |
Hb_008667_040 |
0.2282428342 |
- |
- |
glycerol-3-phosphate transporter family protein [Populus trichocarpa] |
| 11 |
Hb_007229_070 |
0.2306245341 |
- |
- |
PREDICTED: UPF0565 protein C2orf69 [Jatropha curcas] |
| 12 |
Hb_003061_010 |
0.2307192088 |
- |
- |
hypothetical protein SORBIDRAFT_01g039870 [Sorghum bicolor] |
| 13 |
Hb_126112_010 |
0.2312402977 |
- |
- |
PREDICTED: uncharacterized protein LOC105636715 [Jatropha curcas] |
| 14 |
Hb_136765_010 |
0.2314465075 |
- |
- |
PREDICTED: phosphatidylinositol glycan anchor biosynthesis class U protein-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_011161_070 |
0.231720677 |
- |
- |
PREDICTED: kynurenine--oxoglutarate transaminase 1-like isoform X1 [Populus euphratica] |
| 16 |
Hb_009787_010 |
0.2416480026 |
- |
- |
PREDICTED: 10 kDa chaperonin [Jatropha curcas] |
| 17 |
Hb_000302_160 |
0.2430034836 |
- |
- |
PREDICTED: uncharacterized protein LOC105648403 [Jatropha curcas] |
| 18 |
Hb_027472_030 |
0.2440868587 |
- |
- |
RNA helicase 36, partial [Manihot esculenta] |
| 19 |
Hb_011828_050 |
0.2467787942 |
- |
- |
40S ribosomal protein S23, putative [Ricinus communis] |
| 20 |
Hb_082407_010 |
0.2479278219 |
- |
- |
protein farnesyltransferase alpha subunit, putative [Ricinus communis] |